Importing packages and data
[1]:
import glob
import numpy as np
import pandas as pd
from scipy.stats import pearsonr, chi2_contingency
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
from mpl_toolkits.axes_grid1 import make_axes_locatable
from multiprocessing import Pool
import seaborn as sns
from colav.extract_data import *
from biopandas.pdb import PandasPdb
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from umap import UMAP
/Users/asaeed/miniforge3/envs/comp/lib/python3.9/site-packages/umap/distances.py:1063: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/Users/asaeed/miniforge3/envs/comp/lib/python3.9/site-packages/umap/distances.py:1071: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/Users/asaeed/miniforge3/envs/comp/lib/python3.9/site-packages/umap/distances.py:1086: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/Users/asaeed/miniforge3/envs/comp/lib/python3.9/site-packages/umap/umap_.py:660: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
[2]:
# sns.set_palette(sns.color_palette("Dark2"))
[3]:
plt.rcParams["font.size"] = 14
plt.rcParams["axes.labelsize"] = 16
plt.rcParams["axes.labelweight"] = "regular"
plt.rcParams["axes.titlesize"] = 16
plt.rcParams["axes.titleweight"] = "regular"
resnum_bounds = (7, 279)
[4]:
theseus_strucs = sorted(glob.glob("ptp1b_data/ptp1b_theseus_data/*pdb"))
[5]:
# untreated dihedral data processing
untreated_dihedrals, dihedral_strucs = generate_dihedral_matrix(
theseus_strucs,
resnum_bounds,
save=True,
save_prefix="ptp1b_data/ptp1b",
verbose=True,
)
# untreated_dihedrals, dihedral_strucs = load_dihedral_matrix('ptp1b_data/ptp1b_dh_dict.pkl')
untreated_dihedrals_data = np.hstack(
[np.sin(untreated_dihedrals), np.cos(untreated_dihedrals)]
)
Calculating the dihedral angles...
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1a5y_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_1a5y_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1aax_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1bzc_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1bzh_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1bzj_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c83_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c84_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c85_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c86_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c87_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1c88_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ecv_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1een_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1eeo_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1g1f_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1g1g_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1g1h_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1g7f_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1g7g_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1gfy_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1i57_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1jf7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1jf7_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1kak_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1kav_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1l8g_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1lqf_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1lqf_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1lqf_chainC.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1lqf_chainD.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1nl9_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1nny_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1no6_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1nwe_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1nwl_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1nz7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oem_chainX.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oeo_chainX.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_1oeo_chainX.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oes_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oet_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_1oet_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oeu_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_1oeu_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1oev_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_1oev_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ony_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1onz_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1pa1_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ph0_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ptt_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ptu_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1ptv_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1pty_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1pxh_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1pyn_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q1m_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6j_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6m_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6n_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6n_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6p_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6p_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6s_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6s_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6t_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1q6t_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1qxk_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1sug_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1t48_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1t49_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1t4j_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1wax_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_1xbo_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2azr_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2b07_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2b4s_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_2b4s_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2b4s_chainC.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_2b4s_chainC.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2bgd_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_2bgd_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2bge_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cm2_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cm3_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cm3_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cm7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cm8_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cma_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cmb_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cmc_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cne_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cnf_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cng_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cnh_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2cni_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6f_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6t_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6v_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6w_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6y_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f6z_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f70_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2f71_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2fjm_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_2fjm_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2fjm_chainB.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_2fjm_chainB.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2fjn_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2fjn_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2h4g_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2h4k_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2hb1_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2hnp_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2hnq_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2nt7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2nta_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2qbp_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2qbq_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2qbr_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2qbs_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2veu_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2vev_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2vew_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2vex_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2vey_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2zmm_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_2zn7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3a5j_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3a5k_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3cwe_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_3cwe_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3d9c_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3eax_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3eb1_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3eu0_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_3eu0_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3i7z_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3i80_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3qkp_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3qkq_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3sme_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_3zv2_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4bjo_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4bjo_chainB.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4qah_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4qap_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4qbe_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4qbw_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4y14_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_4y14_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_4y14_chainB.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_4y14_chainB.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5k9v_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5k9w_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka0_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka1_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka2_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka3_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka4_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka7_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka8_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5ka9_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5kaa_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5kab_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5kac_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5kad_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5kad_chainB.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_5kad_chainB.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_5t19_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b8e_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b8t_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b8x_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b8z_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b90_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6b95_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6bai_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6cwu_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6cwv_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6ntp_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6ol4_chainA.pdb
Skipping ptp1b_data/ptp1b_theseus_data/theseus_6ol4_chainA.pdb; insufficient atoms!
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6olq_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6olv_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6omy_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6pfw_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6pg0_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6pgt_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6pha_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6phs_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_6pm8_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_7ken_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_7mm1_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0049_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0056_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0058_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0060_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0070_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0071_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0072_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0076_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0077_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0085_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0105_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0111_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0112_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0116_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0118_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0122_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0175_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0180_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0193_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0202_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0205_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0225_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0234_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0241_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0245_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0249_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0288_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0290_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0296_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0323_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0346_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0363_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0375_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0426_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0432_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0441_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0446_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0517_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0518_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0522_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0528_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0538_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0545_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0548_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0559_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0571_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0572_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0573_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0579_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0580_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0589_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0591_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0608_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0623_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0634_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0639_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0645_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0646_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0652_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0654_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0656_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0660_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0676_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0678_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0679_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0710_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0711_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0718_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0720_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0721_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0723_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0725_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0766_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0772_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0812_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0816_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0822_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0829_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0835_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0842_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0845_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0846_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0847_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0876_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0877_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0884_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0891_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0911_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0912_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0914_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0921_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0935_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0953_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0963_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0965_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0972_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0986_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y0994_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1009_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1011_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1043_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1060_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1103_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1125_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1136_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1153_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1163_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1182_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1207_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1242_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1257_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1261_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1264_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1265_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1271_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1288_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1294_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1298_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1302_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1304_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1312_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1314_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1316_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1317_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1318_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1319_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1320_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1327_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1335_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1339_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1343_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1373_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1402_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1417_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1429_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1438_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1442_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1461_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1486_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1492_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1499_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1525_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1527_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1530_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1532_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1548_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1554_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1589_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1594_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1608_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1612_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1613_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1618_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1629_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1646_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1652_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1703_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1710_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1711_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1718_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1733_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1734_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1759_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1763_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1781_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1787_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1791_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1804_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1819_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1827_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1839_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1841_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1842_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1860_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1865_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1868_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1875_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1885_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1886_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1888_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1902_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1904_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1922_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1933_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1938_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1943_chainA.pdb
Attempting to calculate for ptp1b_data/ptp1b_theseus_data/theseus_PTP1B-y1957_chainA.pdb
Saving dh_dict data!
[6]:
# rope dihedral data processing
rope_dihedrals = pd.read_csv("ptp1b_data/filtered-ptp1b.csv")
rope_strucs = np.array([x[:-2] for x in rope_dihedrals.Name.values])
rope_dihedrals.drop(["Unnamed: 0", "Name"], axis=1, inplace=True)
rope_dihedrals = np.deg2rad(rope_dihedrals.values)
rope_dihedrals_data = np.hstack([np.sin(rope_dihedrals), np.cos(rope_dihedrals)])
[7]:
# Dihedral PCA
# untreated
u_pca = PCA(n_components=10)
u_A = u_pca.fit_transform(untreated_dihedrals_data)
# rope
rope_pca = PCA(n_components=10)
rope_A = rope_pca.fit_transform(rope_dihedrals_data)
[8]:
u_pca.explained_variance_ratio_, rope_pca.explained_variance_ratio_
[8]:
(array([0.29747788, 0.06309758, 0.037037 , 0.03535305, 0.0287802 ,
0.02453796, 0.02153294, 0.01868356, 0.01458321, 0.01343809]),
array([0.26460967, 0.05141095, 0.03610283, 0.03250671, 0.02652774,
0.0217012 , 0.02028537, 0.01891694, 0.01596703, 0.01525418]))
[9]:
# pairwise distances data processing
# pw_data, pw_strucs = generate_pw_matrix(theseus_strucs,
# resnum_bounds,
# save=True,
# save_prefix='ptp1b_data/ptp1b',
# verbose=False)
pw_data, pw_strucs = load_pw_matrix("ptp1b_data/ptp1b_pw_dict.pkl")
[10]:
# Pairwise Distance PCA
pw_pca = PCA(n_components=10)
pw_A = pw_pca.fit_transform(pw_data)
[11]:
pw_pca.explained_variance_ratio_
[11]:
array([0.59867585, 0.0718247 , 0.06767347, 0.04182721, 0.03015834,
0.0148717 , 0.01308885, 0.01175772, 0.00932679, 0.00816632])
[12]:
# strain data processing
# sa_data, sa_strucs = generate_strain_matrix(
# theseus_strucs,
# "ptp1b_data/ptp1b_theseus_data/theseus_1nwl_chainA.pdb",
# "sheart",
# resnum_bounds,
# save=True,
# save_prefix="ptp1b_data/ptp1b_1nwl",
# verbose=False,
# )
sa_data, sa_strucs = load_strain_matrix("ptp1b_data/ptp1b_1nwl_sa_sheart_dict.pkl")
[13]:
# from colav.strain_analysis import determine_shared_atoms
# atom_set = determine_shared_atoms(
# theseus_strucs,
# PandasPdb().read_pdb("ptp1b_data/ptp1b_theseus_data/theseus_1nwl_chainA.pdb").df['ATOM'],
# resnum_bounds,
# save=True,
# save_prefix='ptp1b_data/ptp1b'
# )
[14]:
# Strain Analysis PCA
sa_pca = PCA(n_components=10)
sa_A = sa_pca.fit_transform(sa_data)
[15]:
sa_pca.explained_variance_ratio_
[15]:
array([0.58555996, 0.08409118, 0.0470309 , 0.03535379, 0.02284818,
0.0167237 , 0.01463643, 0.0122772 , 0.01073388, 0.00857248])
[16]:
conformation_dict = pickle.load(open("ptp1b_data/ptp1b_conformation_dict.pkl", "rb"))
[17]:
space_group_dict = pickle.load(open("ptp1b_data/ptp1b_sg_dict.pkl", "rb"))
[18]:
# truncated to fit w/ resnum_bounds
ptp1b_ss_dict = {
u"\u03b11'": (7,13),
u"\u03b12'": (15,25),
u'\u03b11': (37,44),
u'\u03b21': (56,59),
u'\u03b22': (69,74),
u'\u03b23': (78,84),
u'\u03b12': (91,102),
u'\u03b24': (106,109),
u'\u03b25': (114,115),
u'\u03b26': (118,119),
u'\u03b27': (133,135),
u'\u03b28': (140,149),
u'\u03b29': (153,162),
u'\u03b210': (167,176),
u'\u03b13': (187,201),
u'\u03b211': (211,214),
u'\u03b14': (220,239),
u'\u03b15': (245,254),
u'\u03b16': (263,279),
}
Main text figures
PCA of COLAV representations
[19]:
# PC space plots
fig, ax = plt.subplots(1, 1, figsize=(7,6), dpi=300)
# ax = ax.flatten()
pca_list = [u_A, pw_A, sa_A]
struc_list_list = [dihedral_strucs, pw_strucs, sa_strucs]
title_list = ["Dihedral Angles", "C\u03b1 Pairwise Distances", "Strain Analysis"]
# enumerate the axes and the list of the PCAs
i = 0
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.grid(alpha=0.4)
# determine conformation color
conformations = list()
for k in struc_list_list[i]:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
cluster1 = ax.scatter(
pca_list[i][conformations == "open_open", 0],
pca_list[i][conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
cluster2 = ax.scatter(
pca_list[i][conformations == "open_closed", 0],
pca_list[i][conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
cluster3 = ax.scatter(
pca_list[i][conformations == "closed_open", 0],
pca_list[i][conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
cluster4 = ax.scatter(
pca_list[i][conformations == "closed_closed", 0],
pca_list[i][conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
for j,struc in enumerate(dihedral_strucs):
if '1nwl' in struc or '4qbw' in struc or '1pxh' in struc or '1sug' in struc:
plt.plot(u_A[j,0], u_A[j,1], 'rx')
ax.set_ylabel("PC2")
ax.set_xlabel("PC1")
ax.set_title(f"PTP-1B {title_list[i]}")
ax.legend(
loc="upper left",
handles=[cluster1, cluster2, cluster3, cluster4],
labels=["open/open", "open/closed", "closed/open", "closed/closed"],
)
plt.tight_layout()
plt.show()
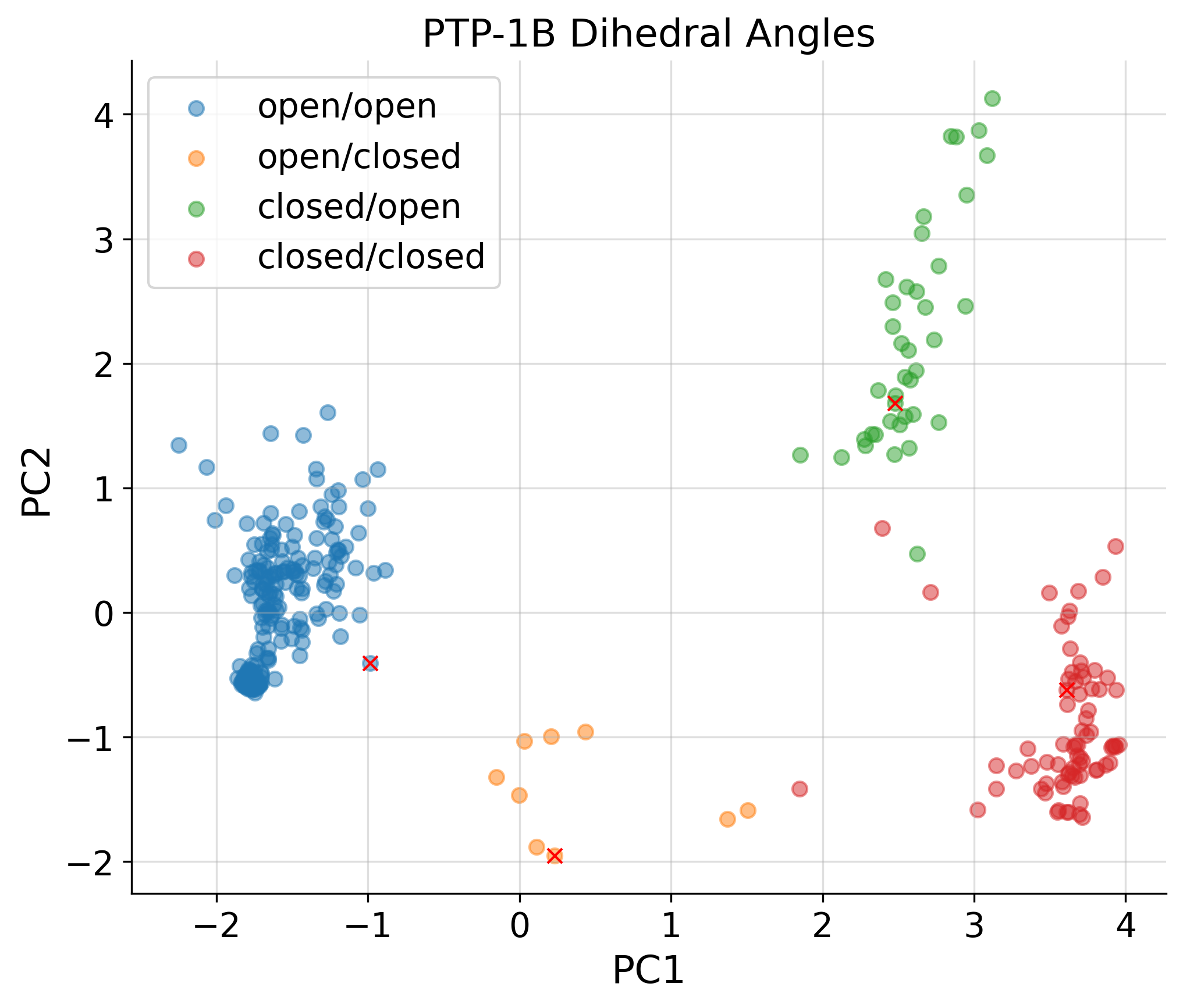
[20]:
# PC space plots
fig, ax = plt.subplots(1, 2, figsize=(14, 6), dpi=300)
ax = ax.flatten()
pca_list = [pw_A, sa_A]
struc_list_list = [pw_strucs, sa_strucs]
title_list = ["C\u03b1 Pairwise Distances", "Strain Analysis"]
# enumerate the axes and the list of the PCAs
for i in np.arange(len(pca_list)):
ax[i].spines['top'].set_visible(False)
ax[i].spines['right'].set_visible(False)
ax[i].grid(alpha=0.4)
# determine conformation color
conformations = list()
for k in struc_list_list[i]:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
if i == 0:
cluster1 = ax[i].scatter(
pca_list[i][conformations == "open_open", 0],
pca_list[i][conformations == "open_open", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
cluster2 = ax[i].scatter(
pca_list[i][conformations == "open_closed", 0],
pca_list[i][conformations == "open_closed", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
cluster3 = ax[i].scatter(
pca_list[i][conformations == "closed_open", 0],
pca_list[i][conformations == "closed_open", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
cluster4 = ax[i].scatter(
pca_list[i][conformations == "closed_closed", 0],
pca_list[i][conformations == "closed_closed", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
ax[i].set_ylabel("PC3")
ax[i].set_xlabel("PC1")
ax[i].set_title(f"PTP-1B {title_list[i]}")
else:
cluster1 = ax[i].scatter(
pca_list[i][conformations == "open_open", 0],
pca_list[i][conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
cluster2 = ax[i].scatter(
pca_list[i][conformations == "open_closed", 0],
pca_list[i][conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
cluster3 = ax[i].scatter(
pca_list[i][conformations == "closed_open", 0],
pca_list[i][conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
cluster4 = ax[i].scatter(
pca_list[i][conformations == "closed_closed", 0],
pca_list[i][conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
ax[i].set_ylabel("PC2")
ax[i].set_xlabel("PC1")
ax[i].set_title(f"PTP-1B {title_list[i]}")
# ax[0].legend(
# loc="upper left",
# handles=[cluster1, cluster2, cluster3, cluster4],
# labels=["open/open", "open/closed", "closed/open", "closed/closed"],
# )
plt.tight_layout()
plt.show()
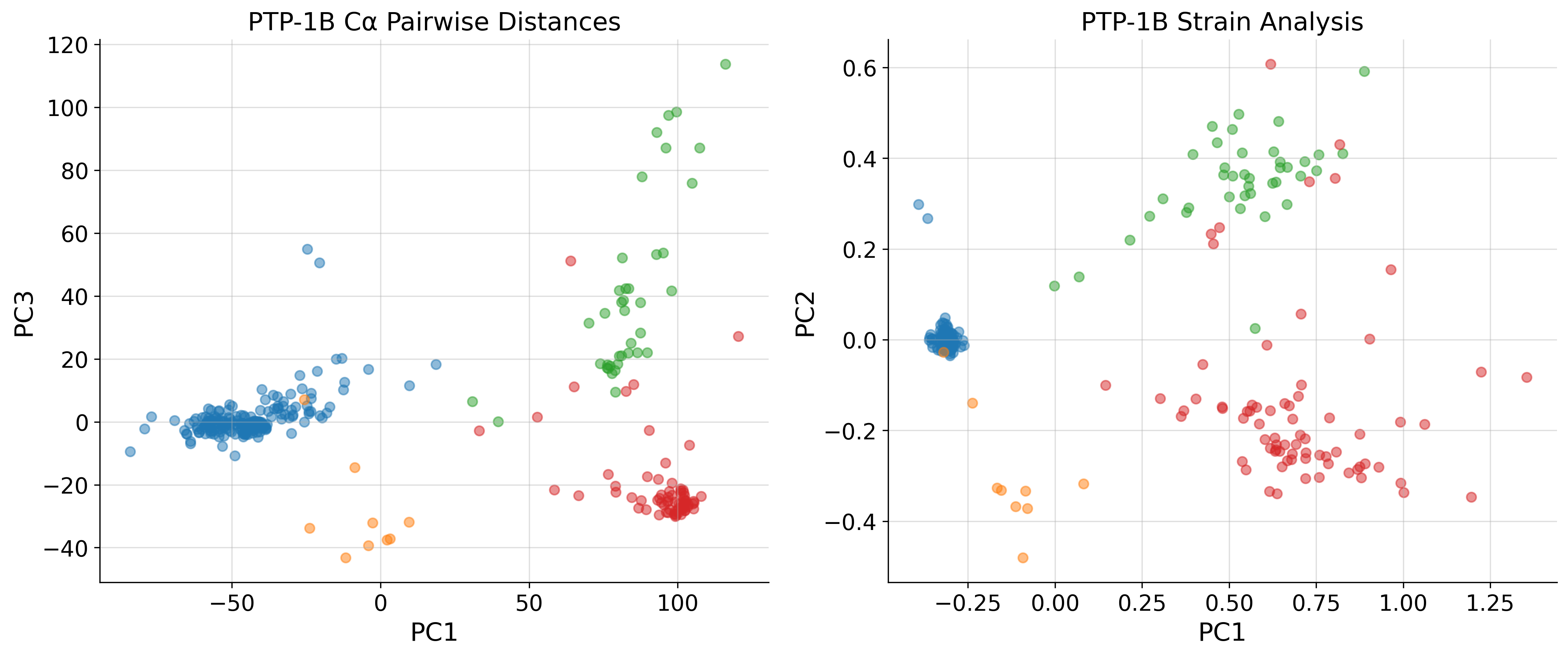
[21]:
# create array for storing correlation coefficients
dh_pw_matrix = list()
dh_sa_matrix = list()
pw_sa_matrix = list()
psi_idx = np.arange(0, untreated_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, untreated_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, untreated_dihedrals_data.shape[1] // 2, 3)
# load the shared atom data
shared_atom_set = pickle.load(open("ptp1b_data/ptp1b_atom_set.pkl", "rb"))
for i in np.arange(10):
for j in np.arange(10):
# calculate the correlation coefficients
dh_pw_matrix.append(
pearsonr(
calculate_dh_rc(u_pca.components_[i, :])[psi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[phi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[omg_idx],
calculate_pw_rc(pw_pca.components_[j, :], resnum_bounds)[1:],
)[0]
)
dh_sa_matrix.append(
pearsonr(
calculate_dh_rc(u_pca.components_[i, :])[psi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[phi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[omg_idx],
calculate_sa_rc(sa_pca.components_[j, :], sorted(shared_atom_set))[1:],
)[0]
)
pw_sa_matrix.append(
pearsonr(
calculate_pw_rc(pw_pca.components_[i, :], resnum_bounds),
calculate_sa_rc(sa_pca.components_[j, :], sorted(shared_atom_set)),
)[0]
)
dh_pw_matrix = np.array(dh_pw_matrix).reshape(10, 10)
dh_sa_matrix = np.array(dh_sa_matrix).reshape(10, 10)
pw_sa_matrix = np.array(pw_sa_matrix).reshape(10, 10)
[22]:
u_pca.explained_variance_ratio_
[22]:
array([0.29747788, 0.06309758, 0.037037 , 0.03535305, 0.0287802 ,
0.02453796, 0.02153294, 0.01868356, 0.01458321, 0.01343809])
[23]:
fig, ax = plt.subplots(1, 3, figsize=(15, 6), dpi=300)
ax = ax.flatten()
axs_list = [("Dihedral Angles", "C\u03b1 Pairwise Distances"), ("Dihedral Angles", "Strain Analysis"), ("C\u03b1 Pairwise\nDistances", "Strain Analysis")]
dot_matrix_list = [dh_pw_matrix, dh_sa_matrix, pw_sa_matrix]
for i in np.arange(3):
im = ax[i].imshow(dot_matrix_list[i][:3,:3], cmap="cividis", interpolation=None)
ax[i].set_xlabel(f"{axs_list[i][1]} RCs")
ax[i].set_ylabel(f"{axs_list[i][0]} RCs")
ax[i].set_title(f"RC Comparison of {axs_list[i][0]} \nand {axs_list[i][1]}" if axs_list[i][0] == "Dihedral Angles" else f"RC Comparison of {axs_list[i][0]} and {axs_list[i][1]}")
ax[i].set_xticks(list(np.arange(3)))
ax[i].set_yticks(list(np.arange(3)))
ax[i].set_xticklabels([f'{k} ({np.round(pw_pca.explained_variance_ratio_[k-1]*100,1)}%)' if i == 0 else f'{k} ({np.round(sa_pca.explained_variance_ratio_[k-1]*100,1)}%)'for k in np.arange(1,4)], rotation=45)
ax[i].set_yticklabels([f'{k} ({np.round(u_pca.explained_variance_ratio_[k-1]*100,1)}%)' if i <= 1 else f'{k} ({np.round(pw_pca.explained_variance_ratio_[k-1]*100,1)}%)'for k in np.arange(1,4)])
divider = make_axes_locatable(ax[i])
cax = divider.append_axes("right", "5%", pad="3%")
plt.colorbar(im, cax=cax)
plt.tight_layout()
plt.show()
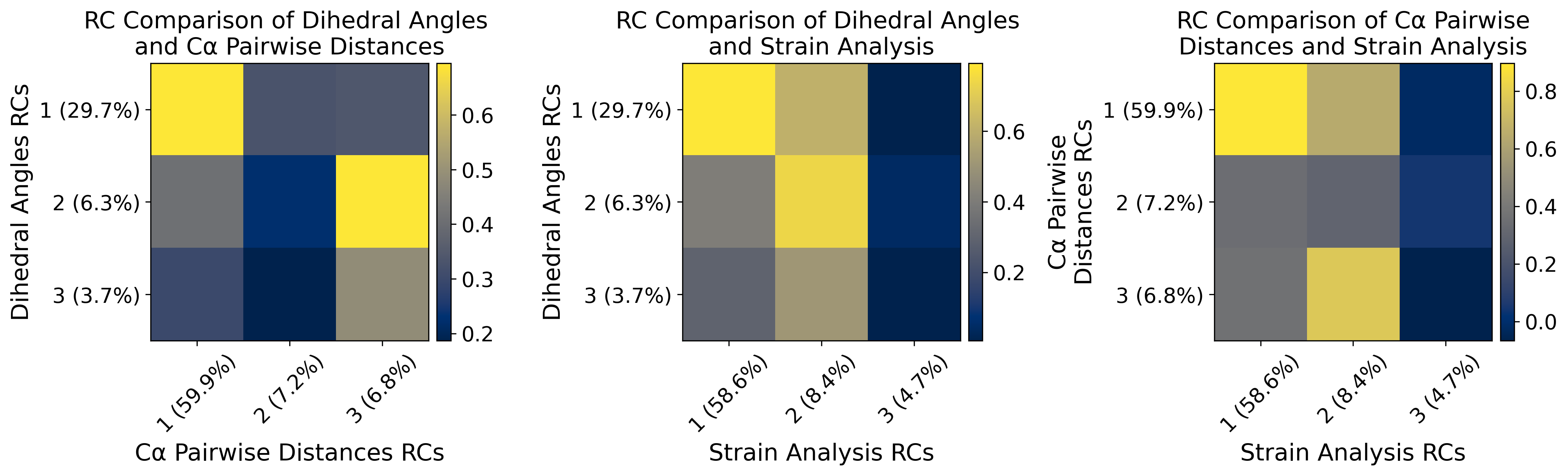
[24]:
fig, ax = plt.subplots(1, 3, figsize=(18, 6), dpi=300)
ax = ax.flatten()
for i in np.arange(3):
dh_tl = calculate_dh_rc(u_pca.components_[i, :])[psi_idx] + calculate_dh_rc(u_pca.components_[i, :])[phi_idx] + calculate_dh_rc(u_pca.components_[i, :])[omg_idx]
pw_tl = calculate_pw_rc(pw_pca.components_[i, :], resnum_bounds)[1:]
sa_tl = calculate_sa_rc(sa_pca.components_[i, :], sorted(shared_atom_set))[1:]
im = ax[i].imshow(np.array([
# [pearsonr(dh_tl, dh_tl)[0], pearsonr(dh_tl, pw_tl)[0], pearsonr(dh_tl, sa_tl)[0]],
# [pearsonr(pw_tl, dh_tl)[0], pearsonr(pw_tl, pw_tl)[0], pearsonr(pw_tl, sa_tl)[0]],
# [pearsonr(sa_tl, dh_tl)[0], pearsonr(sa_tl, pw_tl)[0], pearsonr(sa_tl, sa_tl)[0]],
[0, pearsonr(dh_tl, pw_tl)[0], pearsonr(dh_tl, sa_tl)[0]],
[pearsonr(pw_tl, dh_tl)[0], 0, pearsonr(pw_tl, sa_tl)[0]],
[pearsonr(sa_tl, dh_tl)[0], pearsonr(sa_tl, pw_tl)[0], 0],
]), cmap="cividis", interpolation=None)
# ax[i].set_xlabel(f"Representation")
# ax[i].set_ylabel(f"Representation")
ax[i].set_title(f"RC{i+1} Comparison")
ax[i].set_xticks(list(np.arange(3)))
ax[i].set_xticklabels(['Dihedral\nAngles', u'C\u03b1 Pairwise\nDistances', 'Strain\nAnalysis'])
ax[i].set_yticks(list(np.arange(3)))
ax[i].set_yticklabels(['Dihedral\nAngles', u'C\u03b1 Pairwise\nDistances', 'Strain\nAnalysis'])
divider = make_axes_locatable(ax[i])
cax = divider.append_axes("right", "5%", pad="3%")
plt.colorbar(im, cax=cax)
plt.tight_layout()
plt.show()
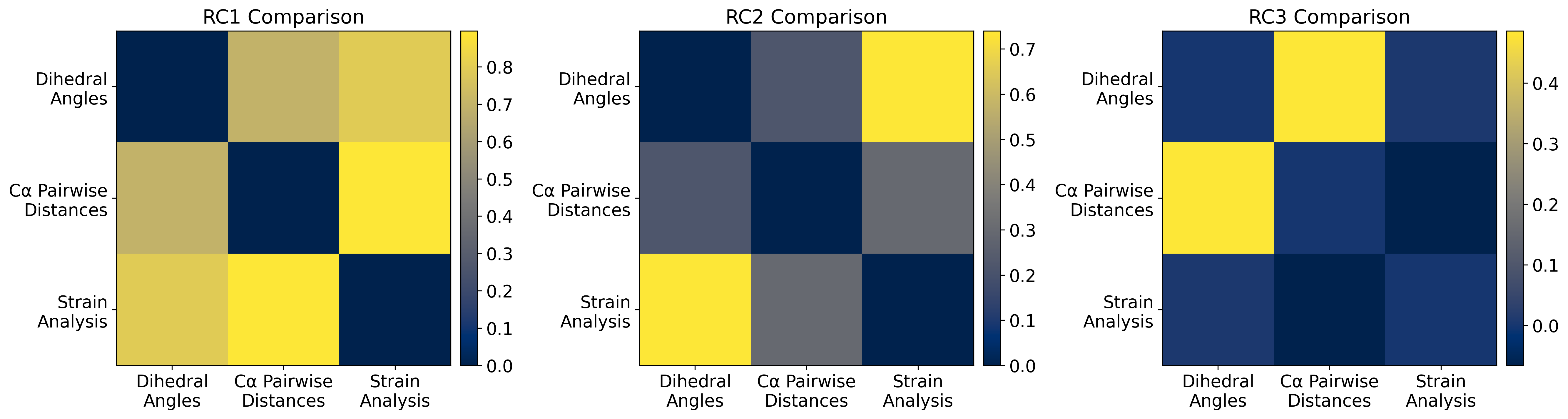
Dihedral Angle PCA space (PC1 & PC2) with RC1 & RC2
[25]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in dihedral_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
u_A[conformations == "open_open", 0],
u_A[conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
u_A[conformations == "open_closed", 0],
u_A[conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
u_A[conformations == "closed_open", 0],
u_A[conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
u_A[conformations == "closed_closed", 0],
u_A[conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
# axd[ax].scatter(
# u_A[[33, 45, 59, 125], 0],
# u_A[[33, 45, 59, 125], 1],
# marker="x",
# s=100,
# c="k",
# label="examples",
# )
axd[ax].set_ylabel("PC2")
axd[ax].set_xlabel("PC1")
axd[ax].set_title("PTP-1B Dihedral Angles")
axd[ax].legend(loc="upper left")
else:
psi_idx = np.arange(0, untreated_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, untreated_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, untreated_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i - 1])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i - 1])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i - 1])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(f"PTP-1B Dihedral Angles RC{i}")
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
# axd[ax].set_yticks(np.arange(0, axd[ax].get_yticks().max(), 0.1), labels=np.round(np.arange(0, axd[ax].get_yticks().max(), 0.1), 2))
axd[ax].add_patch(
Rectangle(
(176, 0),
width=12,
height=3.5, # WPD loop
facecolor="#785EF0",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(237, 0),
width=6,
height=3.5, # L16 loop
facecolor="#FFB000",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
axd[ax].set_ylabel("Relative Value")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
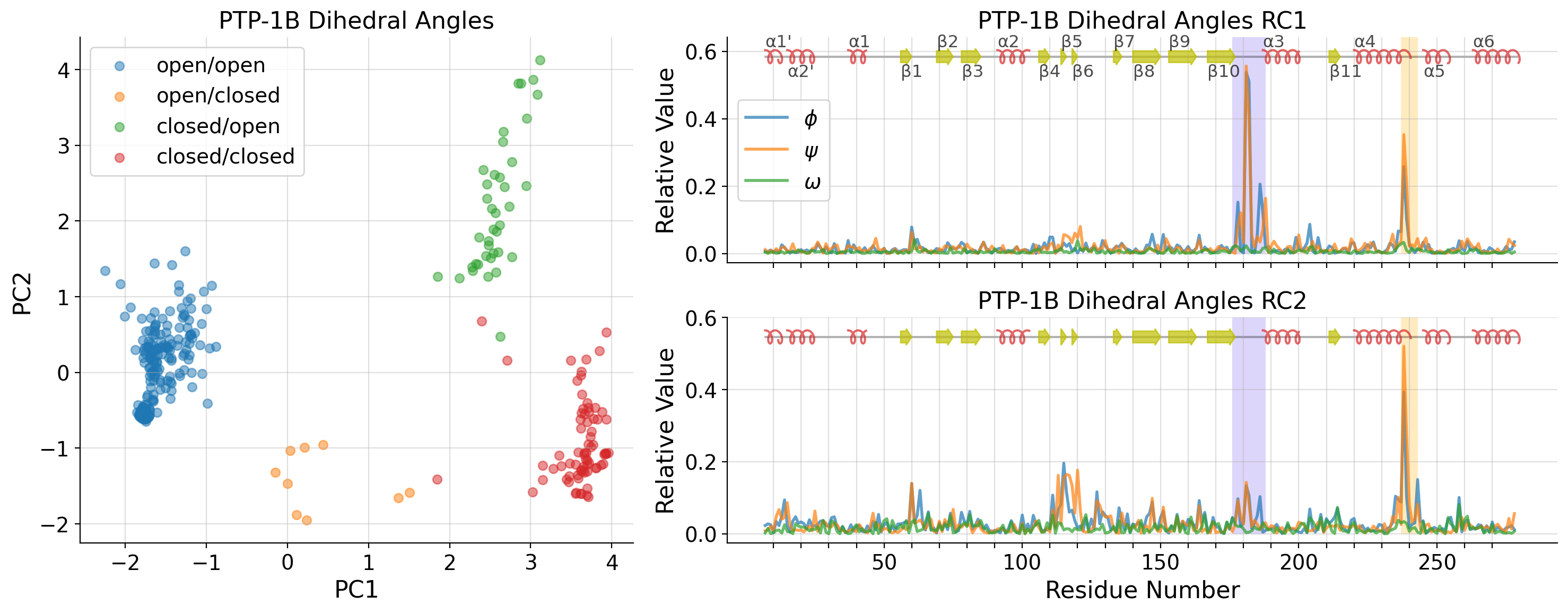
[26]:
l16_loop_dihedrals = untreated_dihedrals[:,687:714]
l16_loop_dihedrals_data = np.hstack([np.sin(l16_loop_dihedrals), np.cos(l16_loop_dihedrals)])
l16_loop_pca = PCA(n_components=10)
l16_loop_A = l16_loop_pca.fit_transform(l16_loop_dihedrals_data)
l16_loop_pca.explained_variance_ratio_
[26]:
array([0.63530741, 0.07368558, 0.06210699, 0.03719484, 0.03084564,
0.02494609, 0.01972787, 0.0180215 , 0.01457365, 0.0123182 ])
[63]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 0, 1]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in dihedral_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
l16_loop_A[conformations == "open_open", 0],
l16_loop_A[conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
l16_loop_A[conformations == "open_closed", 0],
l16_loop_A[conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
l16_loop_A[conformations == "closed_open", 0],
l16_loop_A[conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
l16_loop_A[conformations == "closed_closed", 0],
l16_loop_A[conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
# for j in np.arange(l16_loop_A.shape[0]):
# if l16_loop_A[j,1] > 0 and l16_loop_A[j,0] > -0.5:
# axd[ax].annotate(dihedral_strucs[j].split('/')[-1][8:-4], (l16_loop_A[j,0], l16_loop_A[j,1]))
axd[ax].set_ylabel("PC2")
axd[ax].set_xlabel("PC1")
axd[ax].set_title("PTP-1B (L16 loop) Dihedral Angles")
elif i == 1:
axd[ax].hist(np.array([l16_loop_A[conformations=='open_open',0],
l16_loop_A[conformations=='open_closed',0],
l16_loop_A[conformations=='closed_open',0],
l16_loop_A[conformations=='closed_closed',0]], dtype='object'),
bins=15, density=False, histtype='bar', stacked=True, label=['open/open', 'open/closed', 'closed/open', 'closed/closed'])
axd[ax].set_title('PTP-1B (L16 loop) Dihedral Angles PC1')
axd[ax].set_xlabel('PC1')
axd[ax].set_ylabel('Counts')
else:
psi_idx = np.arange(0, l16_loop_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, l16_loop_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, l16_loop_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(236, 244+1)
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(l16_loop_pca.components_[tl_list[1]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(l16_loop_pca.components_[tl_list[1]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(l16_loop_pca.components_[tl_list[1]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(
f"PTP-1B (L16 loop) Dihedral Angles RC{tl_list[1]+1}"
)
axd[ax].set_xticks(
[dh for dh in dh_range],
labels=[dh for dh in dh_range],
)
axd[ax].set_ylabel("Relative Value")
axd[ax].set_xlabel("Residue Number")
axd["left"].legend(loc="best")
axd["upper right"].legend(loc="best")
axd["lower right"].legend(loc="upper left")
plt.tight_layout()
plt.show()
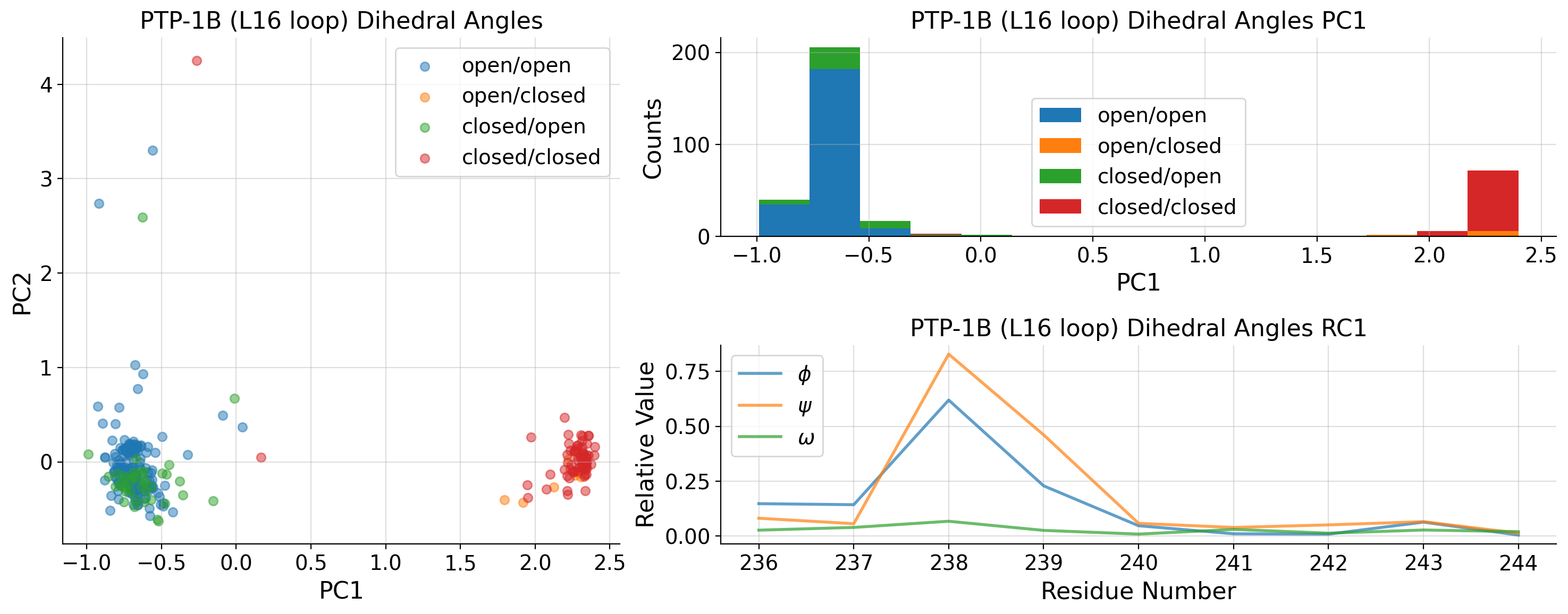
Contingency table and chi-squared test
The null hypothesis of this hypothesis test is that the three regions of PTP-1B – WPD loop, L16 loop, and \alpha7 helix – are
[28]:
a7_helix = list()
for i,k in enumerate(dihedral_strucs):
a7_helix.append(PandasPdb().read_pdb(k).df['ATOM'].residue_number.values[-1])
a7_helix = (np.array(a7_helix) >= 290).astype("int")
[29]:
closed_wpd = np.zeros(len(dihedral_strucs)).astype("int")
closed_l16 = np.zeros(len(dihedral_strucs)).astype("int")
for i,conf in enumerate(conformations):
if conf.startswith("closed"):
closed_wpd[i] += 1
if conf.endswith("closed"):
closed_l16[i] += 1
[30]:
ctable = np.array([[[np.sum(1*(conformations == "open_open") * (1-a7_helix)), np.sum(1*(conformations == "open_closed") * (1-a7_helix))],
[np.sum(1*(conformations == "closed_open") * (1-a7_helix)), np.sum(1*(conformations == "closed_closed") * (1-a7_helix))]],
[[np.sum(1*(conformations == "open_open") * (a7_helix)), np.sum(1*(conformations == "open_closed") * (a7_helix))],
[np.sum(1*(conformations == "closed_open") * (a7_helix)), np.sum(1*(conformations == "closed_closed") * (a7_helix))]]])
[31]:
ctable[0,:,:], ctable[1,:,:]
[31]:
(array([[221, 4],
[ 23, 1]]),
array([[ 7, 5],
[16, 72]]))
[32]:
test1 = chi2_contingency(ctable[0,:,:])
test2 = chi2_contingency(ctable[1,:,:])
test1.statistic + test2.statistic
[32]:
7.480061435105047
[61]:
test1
[61]:
Chi2ContingencyResult(statistic=0.0007653688524590214, pvalue=0.9779290992834958, dof=1, expected_freq=array([[220.48192771, 4.51807229],
[ 23.51807229, 0.48192771]]))
[62]:
test2
[62]:
Chi2ContingencyResult(statistic=7.479296066252588, pvalue=0.006241246758944259, dof=1, expected_freq=array([[ 2.76, 9.24],
[20.24, 67.76]]))
[33]:
test1.pvalue, test2.pvalue
[33]:
(0.9779290992834958, 0.006241246758944259)
[34]:
result = chi2_contingency(ctable)
result
[34]:
Chi2ContingencyResult(statistic=738.115835766452, pvalue=1.942909006865765e-158, dof=4, expected_freq=array([[[129.36241082, 39.72927973],
[ 61.13329119, 18.77501827]],
[[ 51.95277543, 15.95553403],
[ 24.55152257, 7.54016798]]]))
Oxidized conformation of PTP-1B
[67]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 3, 4]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in dihedral_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
u_A[conformations == "open_open", 3],
u_A[conformations == "open_open", 4],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
u_A[conformations == "open_closed", 3],
u_A[conformations == "open_closed", 4],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
u_A[conformations == "closed_open", 3],
u_A[conformations == "closed_open", 4],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
u_A[conformations == "closed_closed", 3],
u_A[conformations == "closed_closed", 4],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
axd[ax].set_ylabel("PC5")
axd[ax].set_xlabel("PC4")
axd[ax].set_title("PTP-1B Dihedral Angles")
axd[ax].legend(loc="upper right")
counter = 0
for i in np.arange(u_A.shape[0]):
if u_A[i,3] > 2 and u_A[i,4] > 2:
if 'PTP1B-y' in dihedral_strucs[i]:
axd[ax].annotate(dihedral_strucs[i].split("/")[-1][14:-11],
(u_A[i,3], u_A[i,4]),
xytext=(u_A[i,3]-0.6 if counter % 2 == 1 else u_A[i,3], u_A[i,4]),
fontsize='small'
)
else:
axd[ax].annotate(dihedral_strucs[i].split("/")[-1][8:-11].upper(),
(u_A[i,3], u_A[i,4]),
xytext=(u_A[i,3]-0.6 if counter % 2 == 0 else u_A[i,3], u_A[i,4]),
fontsize='small'
)
counter += 1
else:
psi_idx = np.arange(0, untreated_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, untreated_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, untreated_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[tl_list[i]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[tl_list[i]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(u_pca.components_[tl_list[i]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(f"PTP-1B Dihedral Angles RC{tl_list[i]+1}")
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
axd[ax].add_patch(
Rectangle(
(213, 0),
width=11,
height=3.5, # P loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(44, 0),
width=7,
height=3.5, # pY loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(60, 0),
width=5,
height=3.5, # Y loop
facecolor="#03c03c",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(257, 0),
width=5,
height=3.5, # Q loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
axd[ax].set_ylabel("Relative Value")
# PDB structures all show the oxidized conformation with sulphenyl-amide ring
# frag screen structures show BME adduct-like conformation
axd["left"].legend(loc="upper left")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
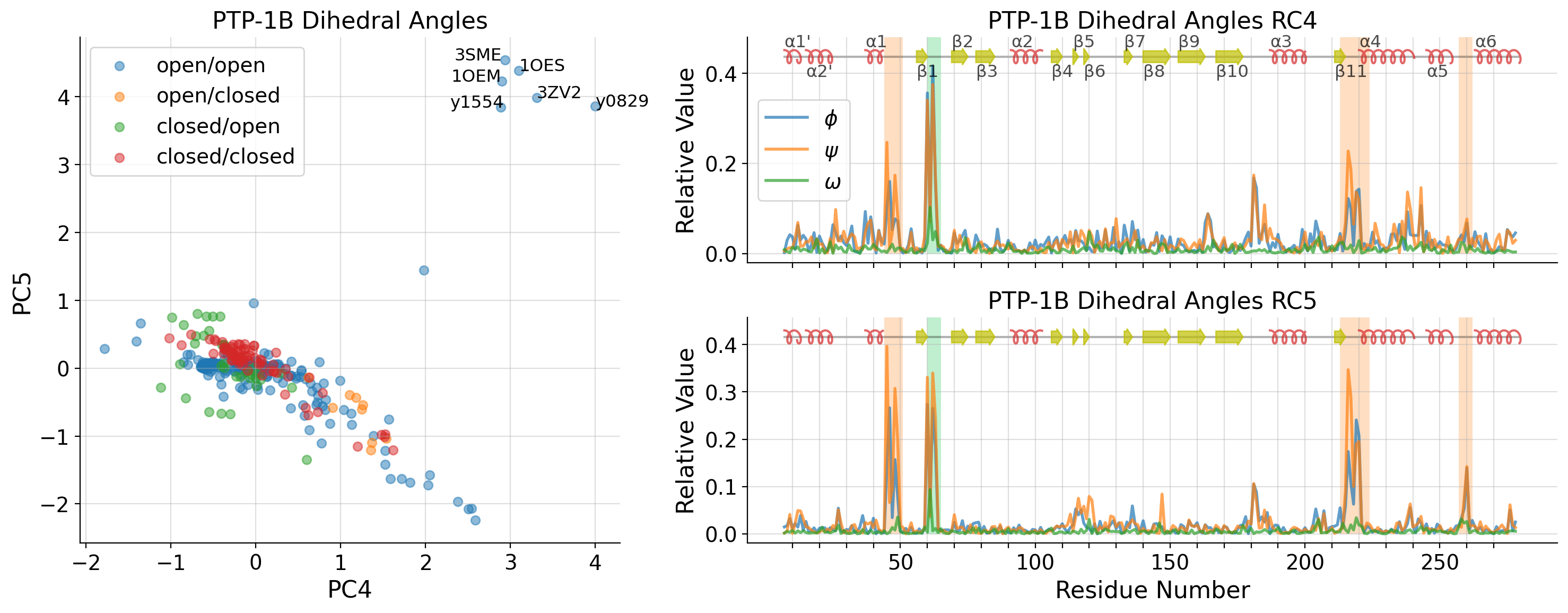
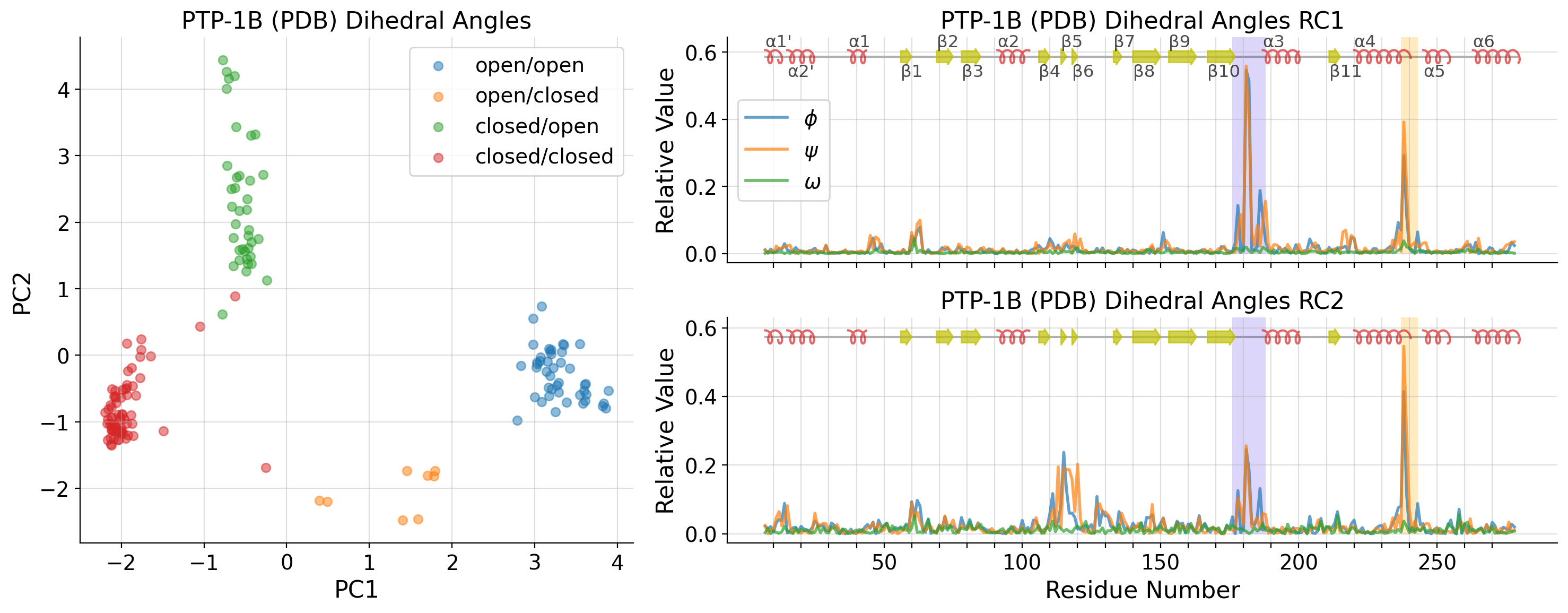
PCA on PDB structures only
[36]:
pdb_strucs = list()
pdb_dihedrals_data = list()
for i, k in enumerate(dihedral_strucs):
if "PTP1B-y" not in k:
pdb_strucs.append(k)
pdb_dihedrals_data.append(untreated_dihedrals_data[i, :])
pdb_dihedrals_data = np.array(pdb_dihedrals_data)
pdb_pca = PCA(n_components=10)
pdb_A = pdb_pca.fit_transform(pdb_dihedrals_data)
[64]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 0, 1]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in pdb_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
pdb_A[conformations == "open_open", 0],
pdb_A[conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
pdb_A[conformations == "open_closed", 0],
pdb_A[conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
pdb_A[conformations == "closed_open", 0],
pdb_A[conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
pdb_A[conformations == "closed_closed", 0],
pdb_A[conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
axd[ax].set_ylabel("PC2")
axd[ax].set_xlabel("PC1")
axd[ax].set_title("PTP-1B (PDB) Dihedral Angles")
else:
psi_idx = np.arange(0, pdb_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, pdb_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, pdb_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(pdb_pca.components_[tl_list[i]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(pdb_pca.components_[tl_list[i]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(pdb_pca.components_[tl_list[i]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(
f"PTP-1B (PDB) Dihedral Angles RC{tl_list[i]+1}"
)
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
axd[ax].add_patch(
Rectangle(
(176, 0),
width=12,
height=3.5, # WPD loop
facecolor="#785EF0",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(237, 0),
width=6,
height=3.5, # L16 loop
facecolor="#FFB000",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
axd[ax].set_ylabel("Relative Value")
axd["left"].legend(loc="best")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
PCA on crystallographic drug fragment screen structures
[38]:
fragment_strucs = list()
fragment_dihedrals_data = list()
for i, k in enumerate(dihedral_strucs):
if "PTP1B-y" in k:
fragment_strucs.append(k)
fragment_dihedrals_data.append(untreated_dihedrals_data[i, :])
fragment_dihedrals_data = np.array(fragment_dihedrals_data)
fragment_pca = PCA(n_components=10)
fragment_A = fragment_pca.fit_transform(fragment_dihedrals_data)
[65]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 4, 6]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in fragment_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
fragment_A[conformations == "open_open", tl_list[1]],
fragment_A[conformations == "open_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
fragment_A[conformations == "open_closed", tl_list[1]],
fragment_A[conformations == "open_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
fragment_A[conformations == "closed_open", tl_list[1]],
fragment_A[conformations == "closed_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
fragment_A[conformations == "closed_closed", tl_list[1]],
fragment_A[conformations == "closed_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
axd[ax].set_ylabel(f"PC{tl_list[2]+1}")
axd[ax].set_xlabel(f"PC{tl_list[1]+1}")
axd[ax].set_title("PTP-1B (Fragment) Dihedral Angles")
axd[ax].legend(loc="upper right")
else:
psi_idx = np.arange(0, fragment_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, fragment_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, fragment_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(
f"PTP-1B (Fragment) Dihedral Angles RC{tl_list[i]+1}"
)
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
axd[ax].add_patch(
Rectangle(
(176, 0),
width=12,
height=3.5, # WPD loop
facecolor="#785EF0",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(237, 0),
width=6,
height=3.5, # L16 loop
facecolor="#FFB000",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
axd[ax].set_ylabel("Relative Value")
axd["left"].legend(loc="best")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
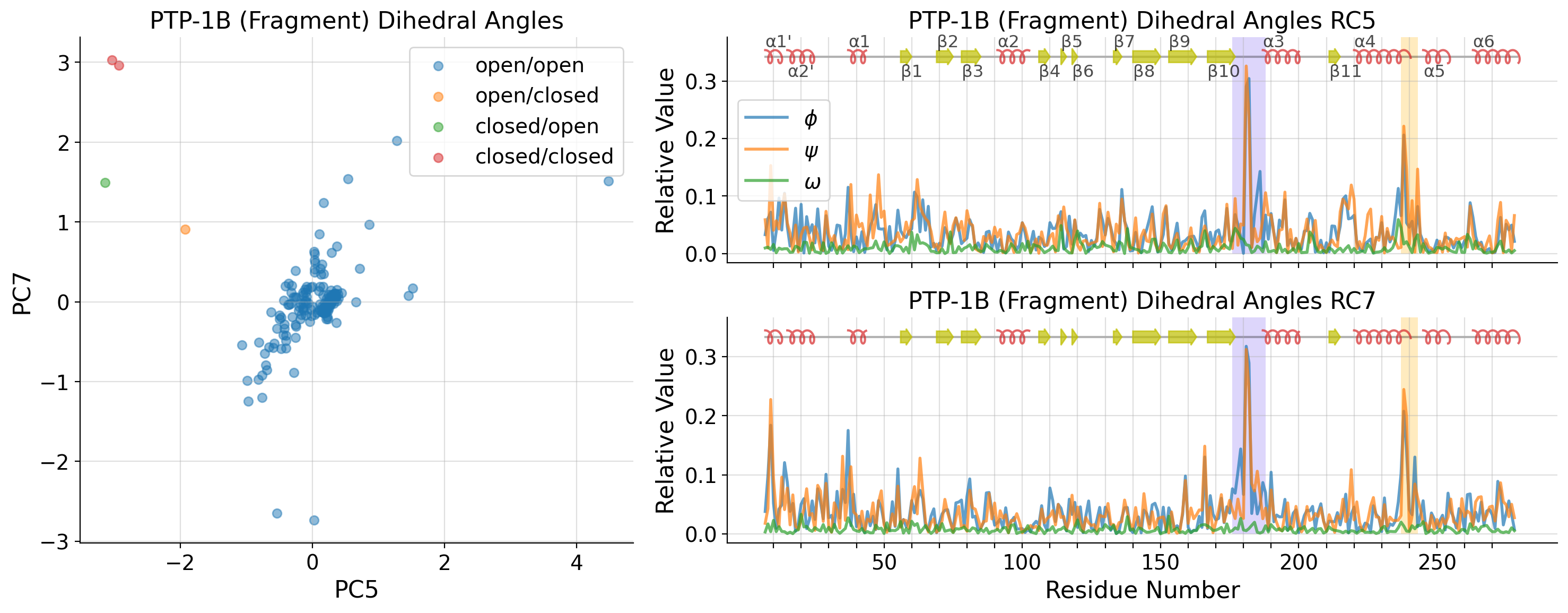
[66]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 1, 2]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in fragment_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
fragment_A[conformations == "open_open", tl_list[1]],
fragment_A[conformations == "open_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
fragment_A[conformations == "open_closed", tl_list[1]],
fragment_A[conformations == "open_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
fragment_A[conformations == "closed_open", tl_list[1]],
fragment_A[conformations == "closed_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
fragment_A[conformations == "closed_closed", tl_list[1]],
fragment_A[conformations == "closed_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
axd[ax].set_ylabel(f"PC{tl_list[2]+1}")
axd[ax].set_xlabel(f"PC{tl_list[1]+1}")
axd[ax].set_title("PTP-1B (Fragment) Dihedral Angles")
axd[ax].legend(loc="upper right")
else:
psi_idx = np.arange(0, fragment_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, fragment_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, fragment_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(
f"PTP-1B (Fragment) Dihedral Angles RC{tl_list[i]+1}"
)
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
axd[ax].add_patch(
Rectangle(
(213, 0),
width=11,
height=3.5, # P loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(44, 0),
width=7,
height=3.5, # pY loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(60, 0),
width=6,
height=3.5, # Y loop
facecolor="slateblue",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(257, 0),
width=5,
height=3.5, # Q loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
# if ax == 'upper right':
# axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
# counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
# if ax == 'upper right':
# axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
# counter += 1
axd[ax].set_ylabel("Relative Value")
axd["left"].legend(loc="best")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
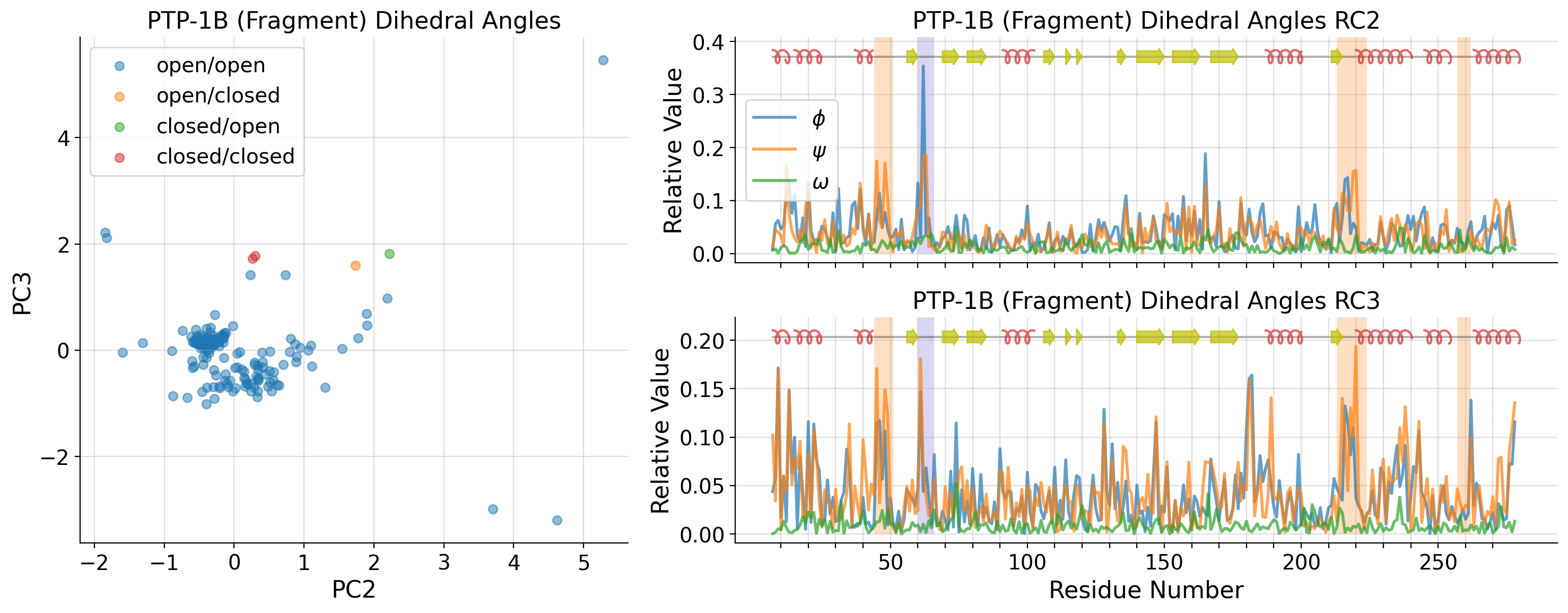
[41]:
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(15, 6),
layout="constrained",
dpi=200,
)
tl_list = [-1, 0, 3]
for i, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if i == 0:
# determine conformation color
conformations = list()
for k in fragment_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
axd[ax].scatter(
fragment_A[conformations == "open_open", tl_list[1]],
fragment_A[conformations == "open_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
axd[ax].scatter(
fragment_A[conformations == "open_closed", tl_list[1]],
fragment_A[conformations == "open_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
axd[ax].scatter(
fragment_A[conformations == "closed_open", tl_list[1]],
fragment_A[conformations == "closed_open", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
axd[ax].scatter(
fragment_A[conformations == "closed_closed", tl_list[1]],
fragment_A[conformations == "closed_closed", tl_list[2]],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
axd[ax].set_ylabel(f"PC{tl_list[2]+1}")
axd[ax].set_xlabel(f"PC{tl_list[1]+1}")
axd[ax].set_title("PTP-1B (Fragment) Dihedral Angles PCA")
axd[ax].legend(loc="upper right")
else:
psi_idx = np.arange(0, fragment_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, fragment_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, fragment_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
(phi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.7,
)
(psi_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.7,
)
(omg_trace,) = axd[ax].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[tl_list[i]])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.7,
)
ylims = axd[ax].get_ylim()
axd[ax].set_title(
f"PTP-1B (Fragment) Dihedral Angles RC{tl_list[i]+1}"
)
axd[ax].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
axd[ax].add_patch(
Rectangle(
(176, 0),
width=12,
height=3.5, # WPD loop
facecolor="#785EF0",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(237, 0),
width=6,
height=3.5, # L16 loop
facecolor="#FFB000",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(213, 0),
width=11,
height=3.5, # P loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(44, 0),
width=7,
height=3.5, # pY loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(60, 0),
width=5,
height=3.5, # Y loop
facecolor="slateblue",
alpha=0.25,
fill=True,
)
)
axd[ax].add_patch(
Rectangle(
(257, 0),
width=5,
height=3.5, # Q loop
facecolor="#ff7f0e",
alpha=0.25,
fill=True,
)
)
axd[ax].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
axd[ax].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
axd[ax].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
axd[ax].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if ax == 'upper right':
axd[ax].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
axd[ax].set_ylabel("Relative Value")
axd["left"].legend(loc="best")
axd["upper right"].set_xticklabels([])
axd["upper right"].legend(loc="center left")
axd["lower right"].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
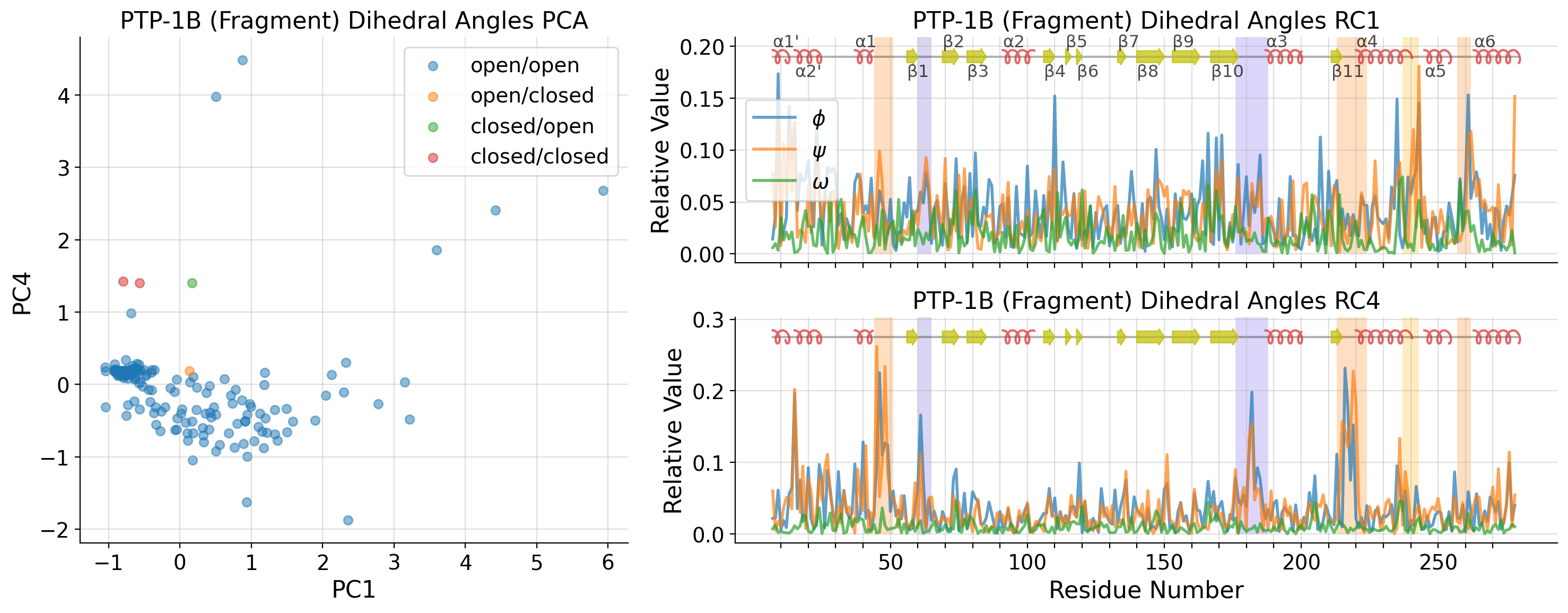
[42]:
fig, ax = plt.subplots(1, 2, figsize=(10, 5), dpi=300)
ax = ax.flatten()
dot_matrix_all = list()
dot_matrix_pdb = list()
for i in np.arange(10):
for j in np.arange(10):
dot_matrix_all.append(
pearsonr(
calculate_dh_rc(u_pca.components_[i, :])[psi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[phi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[omg_idx],
calculate_dh_rc(fragment_pca.components_[j, :])[psi_idx]
+ calculate_dh_rc(fragment_pca.components_[j, :])[phi_idx]
+ calculate_dh_rc(fragment_pca.components_[j, :])[omg_idx]
)[0]
)
dot_matrix_pdb.append(
pearsonr(
calculate_dh_rc(pdb_pca.components_[i, :])[psi_idx]
+ calculate_dh_rc(pdb_pca.components_[i, :])[phi_idx]
+ calculate_dh_rc(pdb_pca.components_[i, :])[omg_idx],
calculate_dh_rc(fragment_pca.components_[j, :])[psi_idx]
+ calculate_dh_rc(fragment_pca.components_[j, :])[phi_idx]
+ calculate_dh_rc(fragment_pca.components_[j, :])[omg_idx],
)[0]
)
dot_matrix_all = np.array(dot_matrix_all).reshape(10, 10)
dot_matrix_pdb = np.array(dot_matrix_pdb).reshape(10, 10)
for i in np.arange(2):
im = ax[i].imshow(dot_matrix_all if i == 0 else dot_matrix_pdb, cmap="cividis", interpolation=None)
divider = make_axes_locatable(ax[i])
cax = divider.append_axes("right", "5%", pad="3%")
plt.colorbar(im, cax=cax)
ax[i].set_xticks(np.arange(10), labels=np.arange(1,11))
ax[i].set_yticks(np.arange(10), labels=np.arange(1,11))
ax[i].set_xlabel("Fragment RCs")
ax[i].set_ylabel("All RCs" if i == 0 else "PDB RCs")
ax[i].set_title("PTP-1B RC Comparison of All\n and Fragment Datasets" if i == 0 else "PTP-1B RC Comparison of PDB\n and Fragment Datasets")
plt.tight_layout()
plt.show()
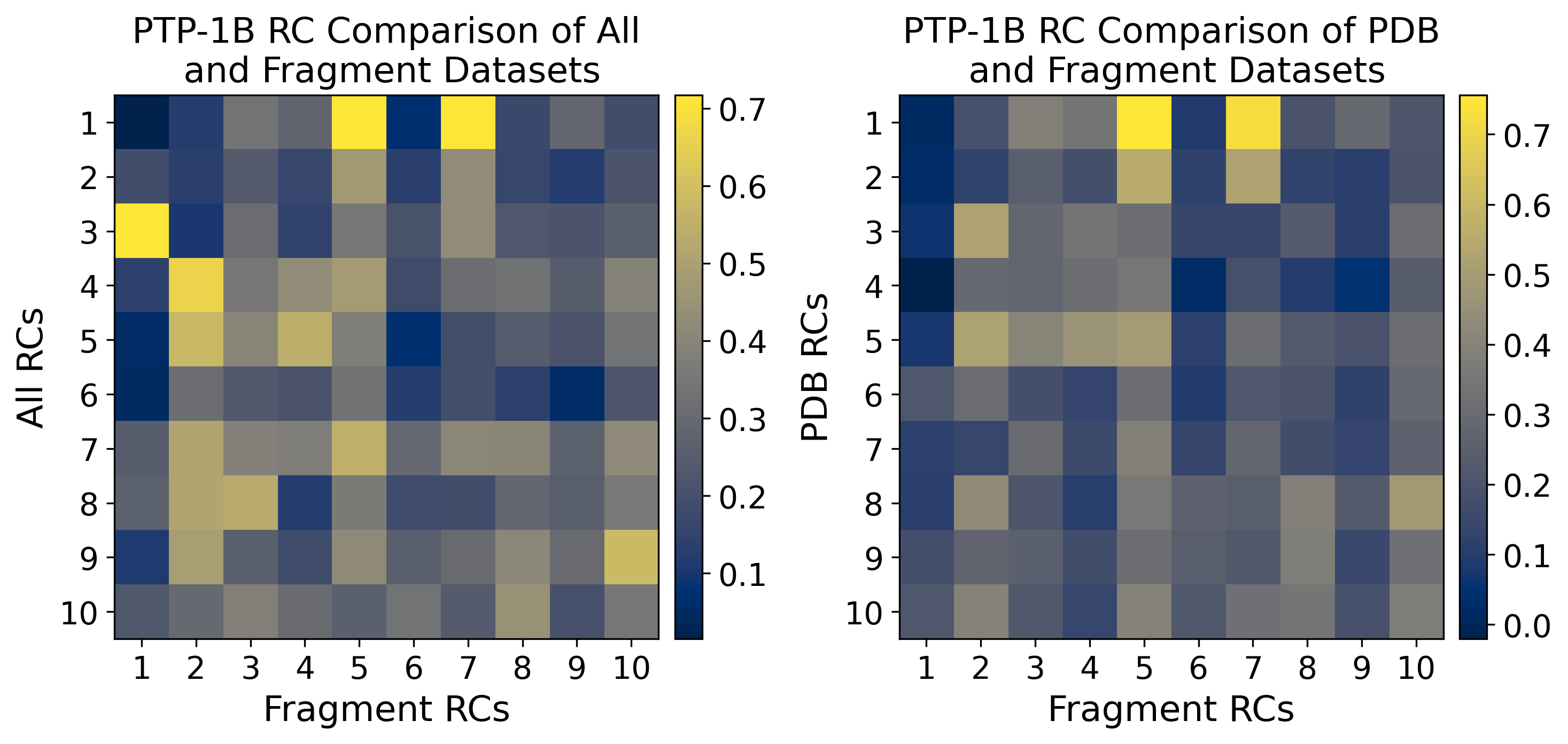
COV and MAT score calculations
[43]:
# calculate_coverage_matching_scores(pdb_strucs, fragment_strucs, (7,279), rmsd_threshold=1.0, verbose=True)
Coverage metric: 96.3%
Matching metric: 0.493
[43]:
(0.9629629629629629, 0.49268775667528564)
[44]:
len(pdb_strucs), len(fragment_strucs)
[44]:
(162, 187)
Conformational changes focusing on individual PCs
[60]:
# allosteric steering
gs_kw = dict(width_ratios=[1, 1.5], height_ratios=[1, 1])
fig, axd = plt.subplot_mosaic(
[["left", "upper right"], ["left", "lower right"]],
gridspec_kw=gs_kw,
figsize=(18, 8),
layout="constrained",
dpi=200,
)
pc1_idxs = np.argsort(u_A[:, 0])
pc2_idxs = np.argsort(u_A[:, 1])
idx_list = [pc1_idxs, pc2_idxs]
for idx, ax in enumerate(axd):
axd[ax].spines['top'].set_visible(False)
axd[ax].spines['right'].set_visible(False)
axd[ax].grid(alpha=0.4)
if idx == 0:
pass
else:
c_dict = {
"open_open": "#1f77b4",
"open_closed": "#ff7f0e",
"closed_open": "#2ca02c",
"closed_closed": "#d62728",
}
conformations = list()
for i in np.array(dihedral_strucs)[idx_list[idx - 1]]:
if i.startswith("ptp1b_data/ptp1b_theseus_data/"):
conformations.append(c_dict[conformation_dict[i.split("/")[-1][8:-4]]])
else:
conformations.append(c_dict[conformation_dict[i]])
conformations = np.array(conformations)
axd[ax].scatter(
np.arange(u_A.shape[0]),
u_A[idx_list[idx - 1], idx - 1],
c=conformations,
alpha=0.5,
marker="o",
edgecolor=None,
)
if idx == 1:
axd[ax].axvline(236.5, linestyle='--', linewidth=2, alpha=0.3, color='r')
axd[ax].text(190, 1, 'Open\nWPD loop', fontsize=16)
axd[ax].text(250, 1, 'Closed\nWPD loop', fontsize=16)
elif idx == 2:
axd[ax].axvline(58.5, linestyle='--', linewidth=2, alpha=0.3, color='r')
axd[ax].text(15, 0, 'Closed\nL16 loop', fontsize=16)
axd[ax].text(70, 0, 'Open\nL16 loop', fontsize=16)
axd[ax].set_ylabel(f"PC{idx} z-score", fontsize=18)
axd[ax].set_title(f"PTP-1B Structures Ordered by PC{idx}", fontsize=20)
axd[ax].set_xlim(left=-5, right=u_A.shape[0] + 5)
axd["upper right"].legend(
loc="upper left",
handles=[cluster1, cluster2, cluster3, cluster4],
labels=["open/open", "open/closed", "closed/open", "closed/closed"],
)
axd["upper right"].set_xticklabels([])
axd["lower right"].set_xlabel("Structures", fontsize=18)
plt.tight_layout()
plt.show()
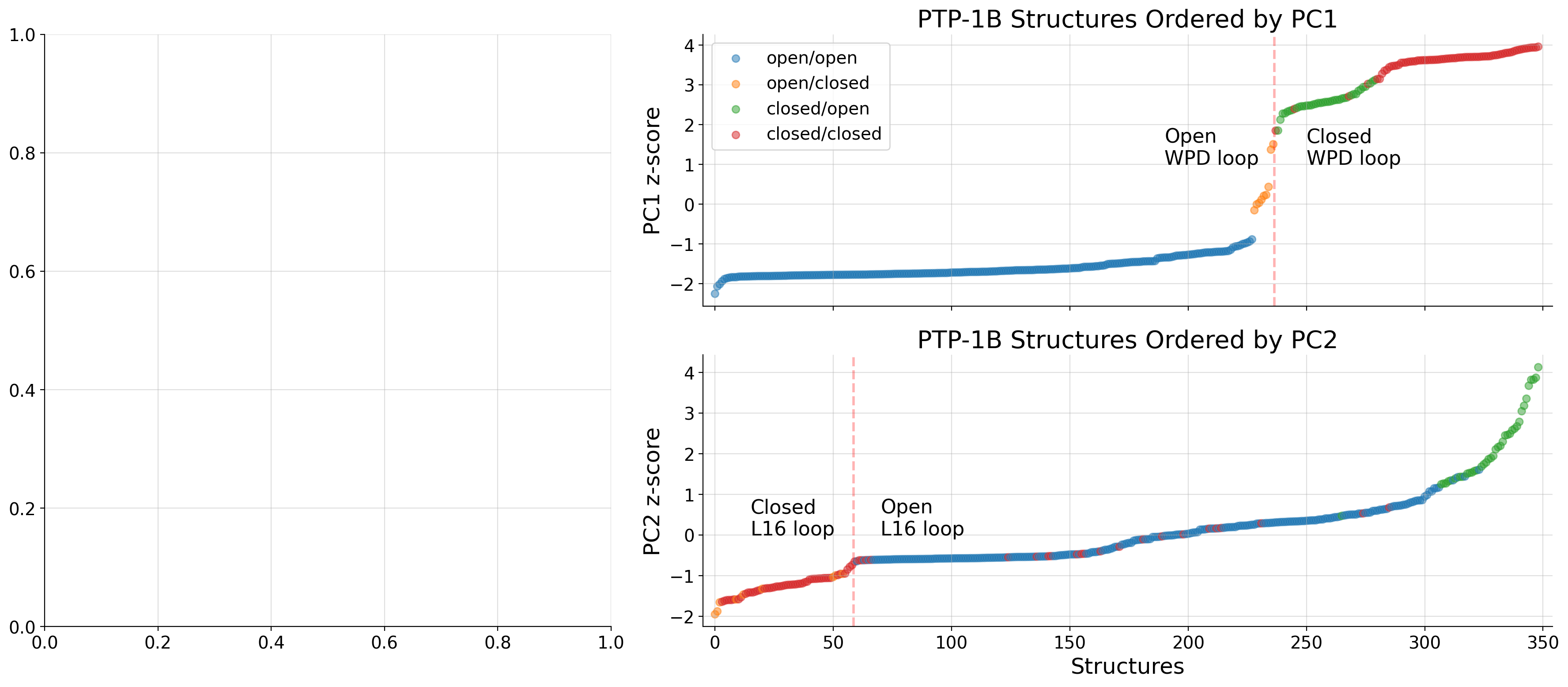
Supplementary Figures
[46]:
# PC space plots
fig, ax = plt.subplots(1, 2, figsize=(15,6), dpi=300)
ax = ax.flatten()
pca_list = [u_A, rope_A, pw_A]
struc_list_list = [dihedral_strucs, rope_strucs, pw_strucs]
title_list = ["Dihedral Angles", "RoPE Dihedral Angles", "C\u03b1 Pairwise Distances"]
# enumerate the axes and the list of the PCAs
for i in np.arange(2):
ax[i].spines['top'].set_visible(False)
ax[i].spines['right'].set_visible(False)
ax[i].grid(alpha=0.4)
# determine conformation color
conformations = list()
for k in struc_list_list[i]:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
cluster1 = ax[i].scatter(
pca_list[i][conformations == "open_open", 0],
pca_list[i][conformations == "open_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
cluster2 = ax[i].scatter(
pca_list[i][conformations == "open_closed", 0],
pca_list[i][conformations == "open_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
cluster3 = ax[i].scatter(
pca_list[i][conformations == "closed_open", 0],
pca_list[i][conformations == "closed_open", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
cluster4 = ax[i].scatter(
pca_list[i][conformations == "closed_closed", 0],
pca_list[i][conformations == "closed_closed", 1],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
ax[i].set_ylabel("PC2")
ax[i].set_xlabel("PC1")
ax[i].set_title("PTP-1B " + title_list[i])
ax[0].legend(
loc="upper left",
handles=[cluster1, cluster2, cluster3, cluster4],
labels=["open/open", "open/closed", "closed/open", "closed/closed"],
)
plt.tight_layout()
plt.show()
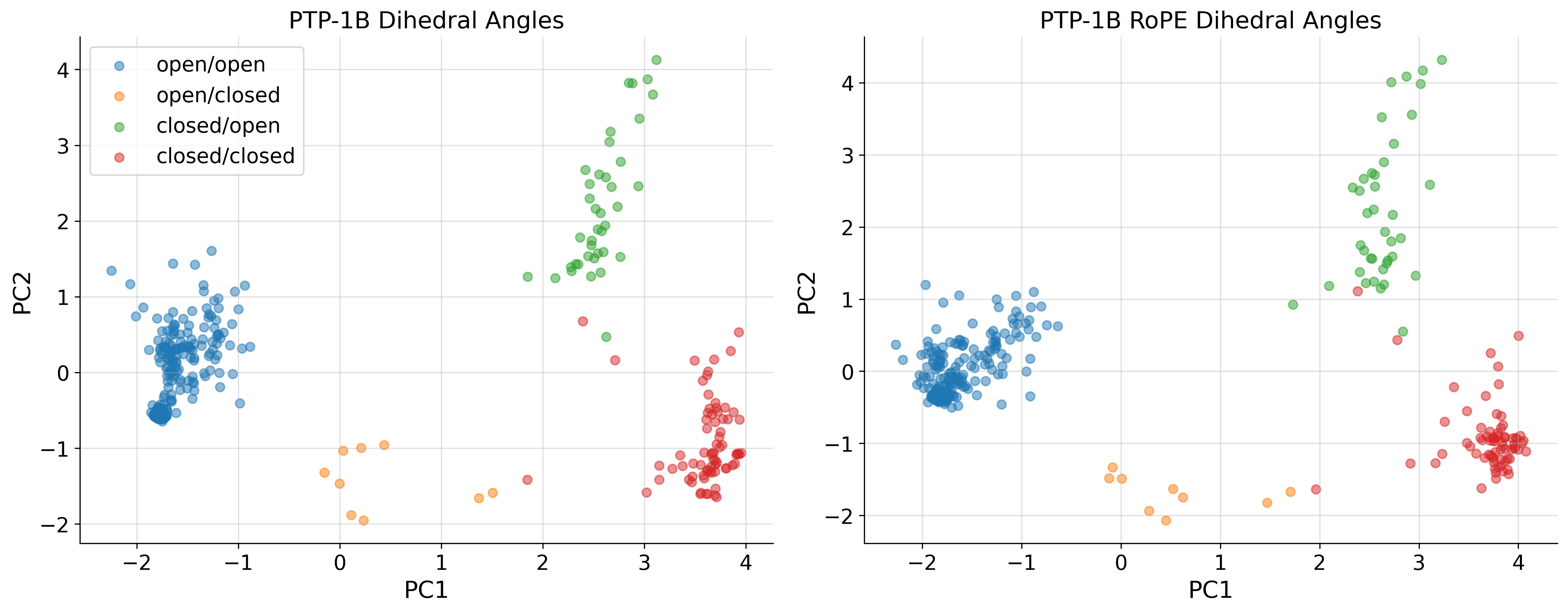
[47]:
fig, ax = plt.subplots(figsize=(10, 5), dpi=300)
# ax = ax.flatten()
dot_matrix_rope = list()
for i in np.arange(10):
for j in np.arange(10):
dot_matrix_rope.append(
pearsonr(
calculate_dh_rc(u_pca.components_[i, :])[psi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[phi_idx]
+ calculate_dh_rc(u_pca.components_[i, :])[omg_idx],
calculate_dh_rc(rope_pca.components_[j, :])[psi_idx]
+ calculate_dh_rc(rope_pca.components_[j, :])[phi_idx]
+ calculate_dh_rc(rope_pca.components_[j, :])[omg_idx],
)[0]
)
dot_matrix_rope = np.array(dot_matrix_rope).reshape(10, 10)
for i in np.arange(2):
im = ax.imshow(dot_matrix_rope, cmap="cividis", interpolation=None)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", "5%", pad="3%")
plt.colorbar(im, cax=cax)
ax.set_xticks(np.arange(10), labels=np.arange(1,11))
ax.set_yticks(np.arange(10), labels=np.arange(1,11))
ax.set_xlabel("RoPE RCs")
ax.set_ylabel("Untreated RCs")
ax.set_title("PTP-1B RC Comparison of\nUntreated and RoPE Structures")
plt.tight_layout()
plt.show()
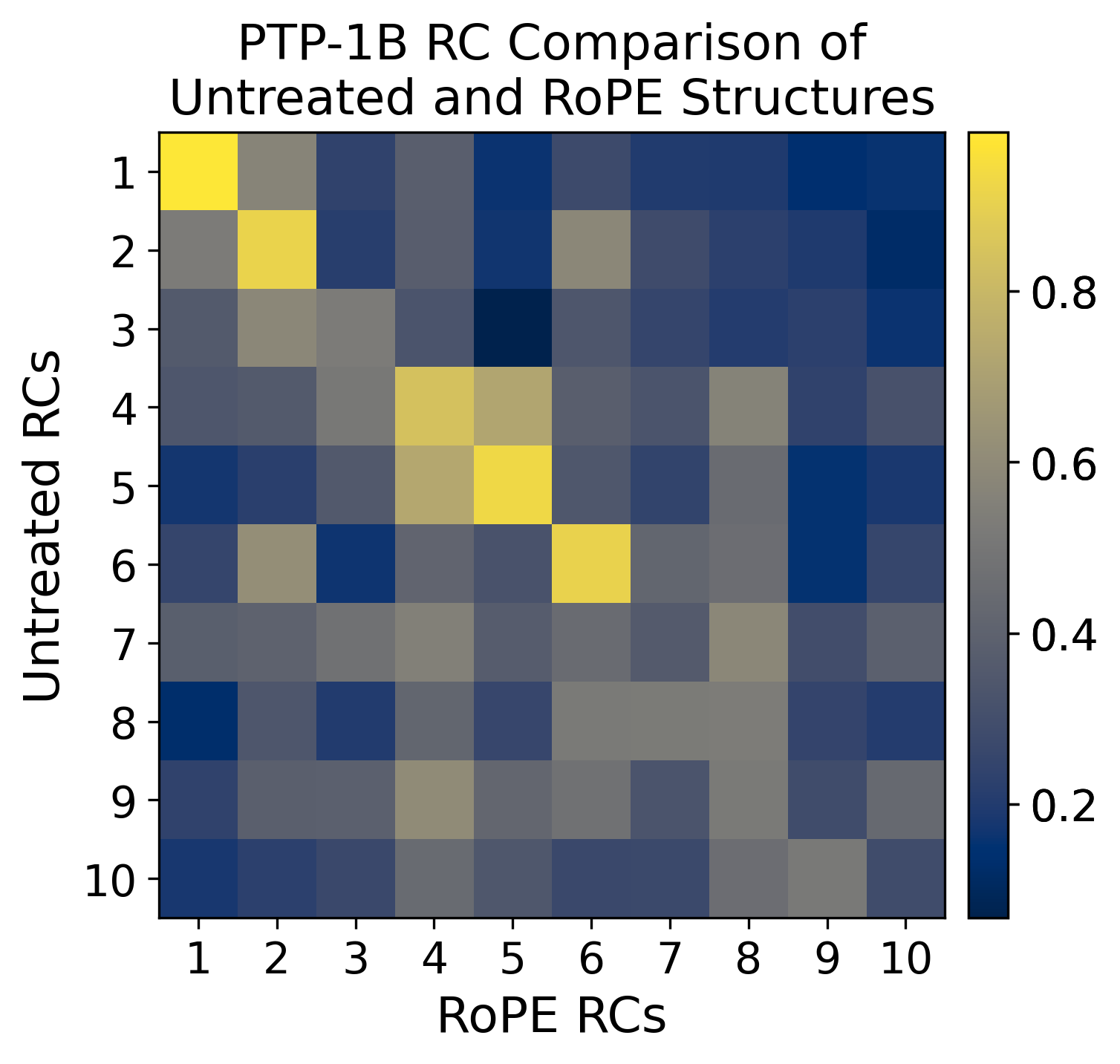
[48]:
# determine the space groups of each of the structures
sgs = list()
for i, struc in enumerate(dihedral_strucs):
sgs.append(space_group_dict[struc.split("/")[-1].split("_")[-2]])
sgs = np.array(sgs)
# PC plot by space group
fig, ax = plt.subplots(1, 1, figsize=(6, 6), dpi=300) # ; ax = ax.flatten()
# for k in np.arange(1):
for i, sg in enumerate(np.unique(sgs)):
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.grid(alpha=0.4)
ax.scatter(
u_A[sgs == sg, 0],
u_A[sgs == sg, 1],
marker="o",
edgecolor=None,
label=f"{sg} ({(sgs== sg).sum()})",
alpha=0.5,
)
ax.set_xlabel("PC1")
ax.set_ylabel("PC2")
ax.set_title("PTP-1B Dihedral Angles by Space Group")
ax.legend()
plt.tight_layout()
plt.show()
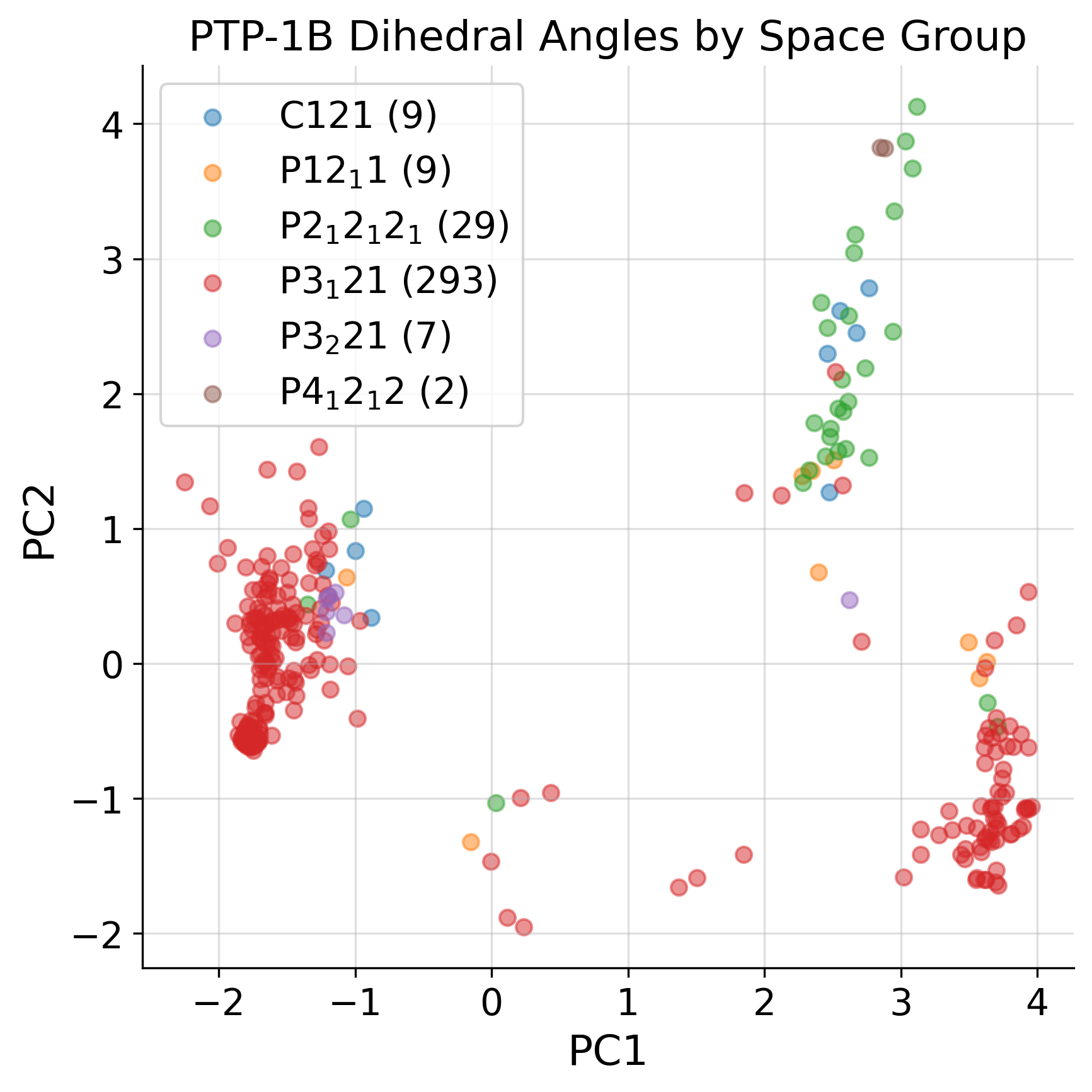
[49]:
conf_list = list()
sg_list = list()
for i,k in enumerate(dihedral_strucs):
sg_list.append(space_group_dict[k.split("/")[-1].split("_")[-2]])
conf_list.append(conformation_dict[k.split("/")[-1][8:-4]])
sg_unq = np.unique(sg_list, return_counts=True, return_inverse=True)
conf_unq = np.unique(conf_list, return_counts=True, return_inverse=True)
ctable = np.zeros((4,6))
for j in np.arange(len(dihedral_strucs)):
ctable[conf_unq[1][j],sg_unq[1][j]] += 1
ctable
[49]:
array([[ 0., 4., 2., 67., 0., 0.],
[ 5., 3., 24., 4., 1., 2.],
[ 0., 1., 1., 7., 0., 0.],
[ 4., 1., 2., 215., 6., 0.]])
[50]:
chi2_contingency(ctable).pvalue
[50]:
2.3184328298516617e-40
[72]:
# calculate t-SNE and UMAP
random_state = 0
u_tsne = TSNE(random_state=random_state, init='random', learning_rate='auto')
u_umap = UMAP(n_neighbors=50, random_state=random_state, init='random')
u_tsne_transform = u_tsne.fit_transform(untreated_dihedrals_data)
u_umap_transform = u_umap.fit_transform(untreated_dihedrals_data)
pw_tsne = TSNE(random_state=random_state, init='random', learning_rate='auto')
pw_umap = UMAP(n_neighbors=50, random_state=random_state, init='random')
pw_tsne_transform = pw_tsne.fit_transform(pw_data)
pw_umap_transform = pw_umap.fit_transform(pw_data)
sa_tsne = TSNE(random_state=random_state, init='random', learning_rate='auto')
sa_umap = UMAP(n_neighbors=50, random_state=random_state, init='random')
sa_tsne_transform = sa_tsne.fit_transform(sa_data)
sa_umap_transform = sa_umap.fit_transform(sa_data)
[73]:
fig, ax = plt.subplots(3, 3, figsize=(18, 18), dpi=200)
ax = ax.flatten()
transform_list = [
u_A,
u_tsne_transform,
u_umap_transform,
pw_A,
pw_tsne_transform,
pw_umap_transform,
sa_A,
sa_tsne_transform,
sa_umap_transform,
]
struc_list_list = [
dihedral_strucs,
dihedral_strucs,
dihedral_strucs,
pw_strucs,
pw_strucs,
pw_strucs,
sa_strucs,
sa_strucs,
sa_strucs,
]
title_list = [
"Dihedral Angles PCA",
"Dihedral Angles t-SNE",
"Dihedral Angles UMAP",
"C\u03b1 Pairwise Distances PCA",
"C\u03b1 Pairwise Distances t-SNE",
"C\u03b1 Pairwise Distances UMAP",
"Strain Analysis PCA",
"Strain Analysis t-SNE",
"Strain Analysis UMAP",
]
for i in np.arange(len(transform_list)):
ax[i].spines['top'].set_visible(False)
ax[i].spines['right'].set_visible(False)
ax[i].grid(alpha=0.4)
# determine conformation color
conformations = list()
for k in struc_list_list[i]:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
cluster1 = ax[i].scatter(
transform_list[i][conformations == "open_open", 0],
transform_list[i][conformations == "open_open", 1] if i != 3 else transform_list[i][conformations == "open_open", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/open",
)
cluster2 = ax[i].scatter(
transform_list[i][conformations == "open_closed", 0],
transform_list[i][conformations == "open_closed", 1] if i != 3 else transform_list[i][conformations == "open_closed", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="open/closed",
)
cluster3 = ax[i].scatter(
transform_list[i][conformations == "closed_open", 0],
transform_list[i][conformations == "closed_open", 1] if i != 3 else transform_list[i][conformations == "closed_open", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/open",
)
cluster4 = ax[i].scatter(
transform_list[i][conformations == "closed_closed", 0],
transform_list[i][conformations == "closed_closed", 1] if i != 3 else transform_list[i][conformations == "closed_closed", 2],
marker="o",
alpha=0.5,
edgecolor=None,
linewidth=1,
label="closed/closed",
)
ax[i].set_ylabel("Coordinate 2")
ax[i].set_xlabel("Coordinate 1")
ax[i].set_title("PTP-1B " + title_list[i])
ax[0].legend(
loc="upper left",
handles=[cluster1, cluster2, cluster3, cluster4],
labels=["open/open", "open/closed", "closed/open", "closed/closed"],
)
plt.tight_layout()
plt.show()
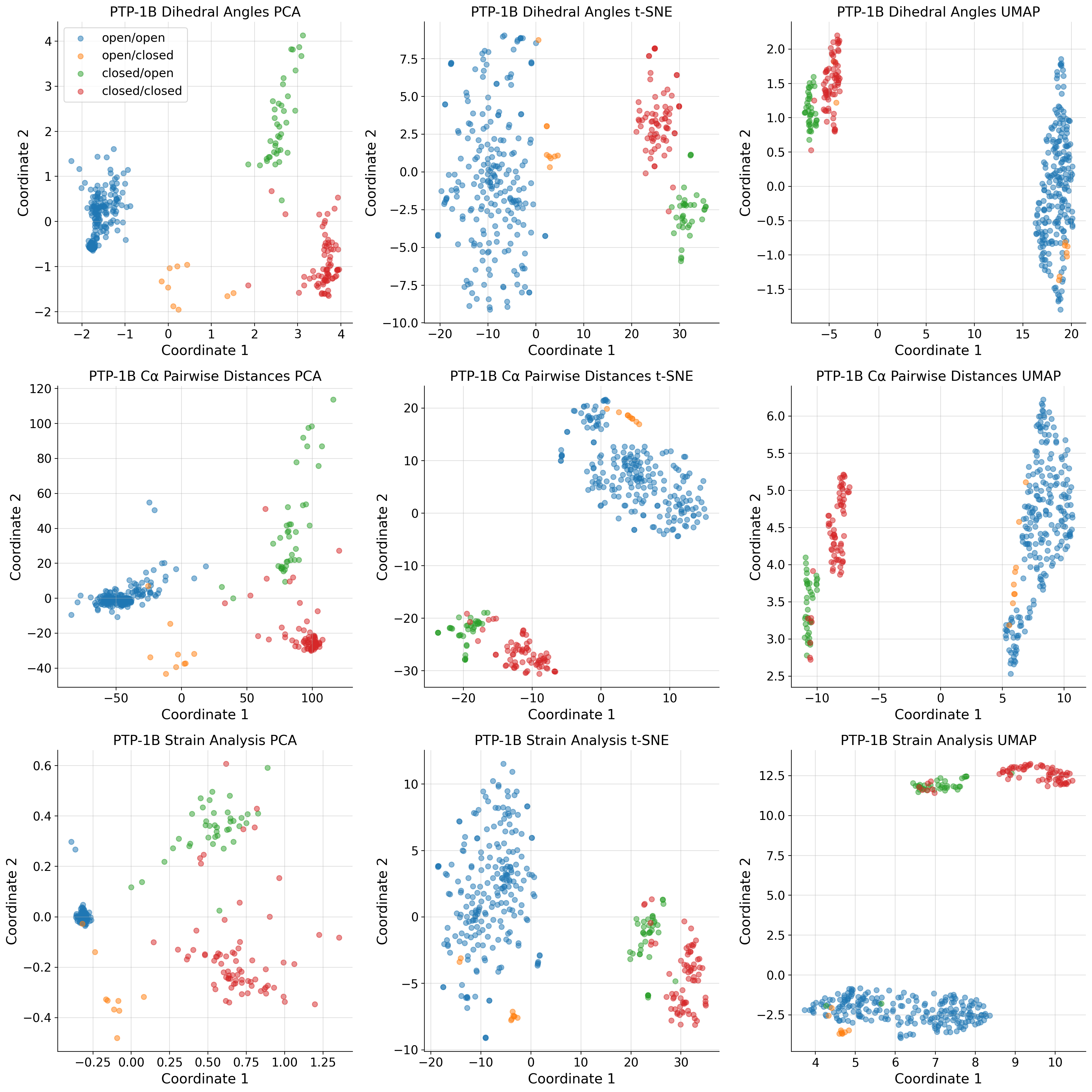
[53]:
fig, ax = plt.subplots(1, 3, figsize=(18, 6), dpi=300)
ax = ax.flatten()
axs_list = [("Dihedral Angles", "C\u03b1 Pairwise Distances"), ("Dihedral Angles", "Strain Analysis"), ("C\u03b1 Pairwise Distances", "Strain Analysis")]
dot_matrix_list = [dh_pw_matrix, dh_sa_matrix, pw_sa_matrix]
for i in np.arange(3):
im = ax[i].imshow(dot_matrix_list[i], cmap="cividis", interpolation=None)
ax[i].set_xlabel(f"{axs_list[i][1]} RCs")
ax[i].set_ylabel(f"{axs_list[i][0]} RCs")
ax[i].set_title(f"RC Comparison of {axs_list[i][0]} \nand {axs_list[i][1]}")
ax[i].set_xticks(np.arange(10), labels=np.arange(1,11))
ax[i].set_yticks(np.arange(10), labels=np.arange(1,11))
divider = make_axes_locatable(ax[i])
cax = divider.append_axes("right", "5%", pad="3%")
plt.colorbar(im, cax=cax)
plt.tight_layout()
plt.show()
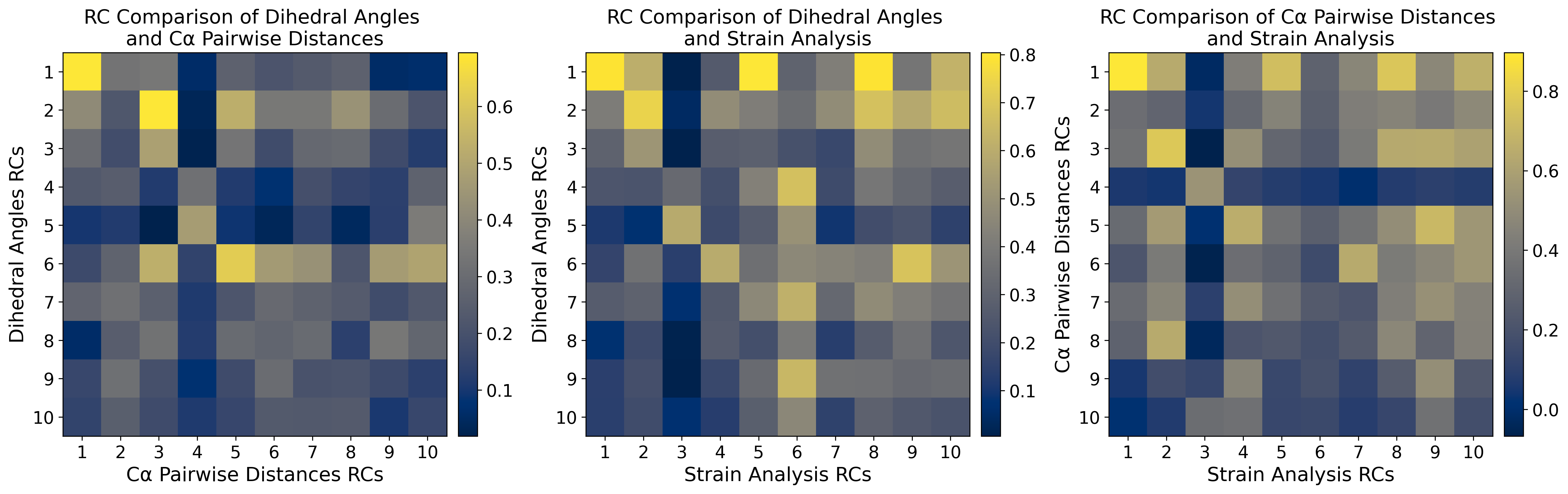
[54]:
dh_sa_matrix[0,0], dh_sa_matrix[1,1]
[54]:
(0.7930733134139794, 0.7402361991988899)
[55]:
# dihedral angle transformed loadings
r = 10
r_list = [r - 1, r - 2]
fig, ax = plt.subplots(r // 2, 2, figsize=(12, 12), dpi=200)
ax = ax.flatten()
psi_idx = np.arange(0, untreated_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, untreated_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, untreated_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
for i in np.arange(r):
ax[i].spines['top'].set_visible(False)
ax[i].spines['right'].set_visible(False)
ax[i].grid(alpha=0.4)
if i % 2 == 0:
(phi_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i//2])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.5,
)
(psi_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i//2])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.5,
)
(omg_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(u_pca.components_[i//2])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.5,
)
ylims = ax[i].get_ylim()
ax[i].set_title(f"PTP-1B Dihedral Angles RC{i//2+1}")
ax[i].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
ax[i].set_ylabel("Relative Value")
if i not in r_list:
ax[i].set_xticklabels([])
ax[i].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
ax[i].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
ax[i].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if i == 0:
ax[i].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
ax[i].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if i == 0:
ax[i].text(v[0], ylims[1]+(1.5*ylims[1]/30) if counter%2 == 0 else ylims[1]-(3*ylims[1]/30), k, fontsize='small', alpha=0.7)
counter += 1
else:
(phi_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[i//2])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.5,
)
(psi_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[i//2])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.5,
)
(omg_trace,) = ax[i].plot(
dh_range,
calculate_dh_rc(fragment_pca.components_[i//2])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.5,
)
ylims = ax[i].get_ylim()
ax[i].set_title(f"Fragment PTP-1B Dihedral Angles RC{i//2+1}")
ax[i].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
ax[i].set_ylabel("Relative Value")
if i not in r_list:
ax[i].set_xticklabels([])
ax[i].plot(np.arange(7,280), [ylims[1]]*273, alpha=0.3, color='k', linestyle='-', zorder=1)
ax[i].set_ylim((ylims[0], ylims[1]*1.1))
counter = 0
for k,v in ptp1b_ss_dict.items():
if k.startswith(u'\u03b1'):
ts = np.linspace(v[0], v[1], num=1000)
ax[i].plot(ts + 1.5*np.sin((ts-v[0])*2*np.pi/3.6), (ylims[1]/30)*np.cos((ts-v[0])*2*np.pi/3.6)+ylims[1], color='tab:red', alpha=0.7, zorder=2)
if i == 0:
ax[i].text(v[0], ylims[1]+0.14 if counter%2 == 0 else ylims[1]+0.03, k, fontsize='small')
counter += 1
else:
ax[i].arrow(v[0],ylims[1],v[1]-v[0]+1,0, width=(ylims[1]/17), head_width=(ylims[1]/12), head_length=2, ec='y', fc='y', length_includes_head=True, alpha=0.7, zorder=3)
if i == 0:
ax[i].text(v[0], ylims[1]+0.14 if counter%2 == 0 else ylims[1]+0.03, k, fontsize='small')
counter += 1
ax[0].legend(loc="center left")
ax[-1].set_xlabel("Residue Number")
ax[-2].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
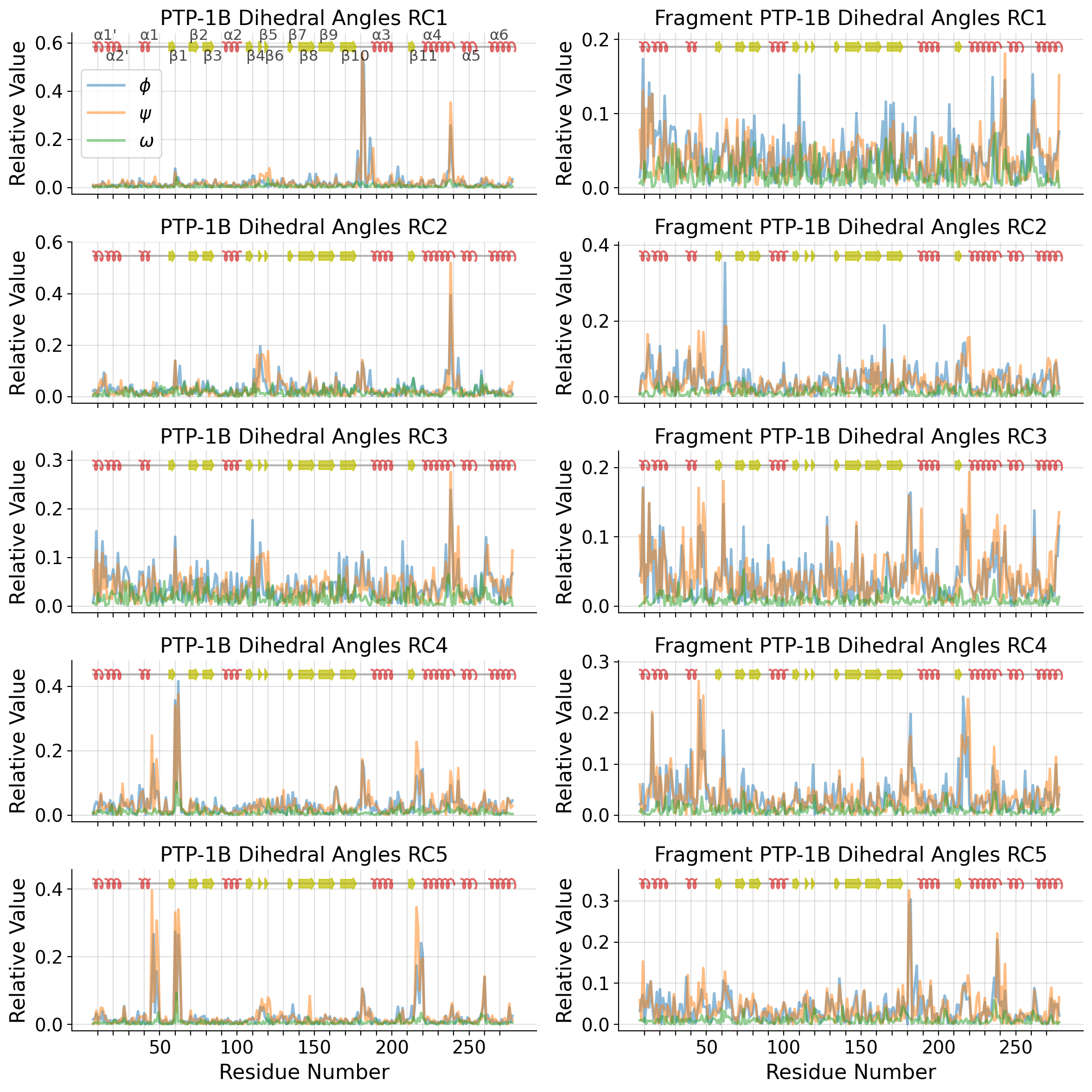
[56]:
# dihedral angle transformed loadings
r = 10
r_list = [r - 1, r - 2]
fig, ax = plt.subplots(r // 2, 2, figsize=(15, 15), dpi=200)
ax = ax.flatten()
psi_idx = np.arange(0, untreated_dihedrals_data.shape[1] // 2, 3)
omg_idx = np.arange(1, untreated_dihedrals_data.shape[1] // 2, 3)
phi_idx = np.arange(2, untreated_dihedrals_data.shape[1] // 2, 3)
dh_range = np.arange(resnum_bounds[0], resnum_bounds[1])
for k in np.arange(r):
(phi_trace,) = ax[k].plot(
dh_range,
calculate_dh_rc(u_pca.components_[k])[phi_idx],
"-",
label=r"$\phi$",
linewidth=2,
alpha=0.5,
)
(psi_trace,) = ax[k].plot(
dh_range,
calculate_dh_rc(u_pca.components_[k])[psi_idx],
"-",
label=r"$\psi$",
linewidth=2,
alpha=0.5,
)
(omg_trace,) = ax[k].plot(
dh_range,
calculate_dh_rc(u_pca.components_[k])[omg_idx],
"-",
label=r"$\omega$",
linewidth=2,
alpha=0.5,
)
ax[k].set_title(f"PTP-1B Dihedral Angles Residue Contribution {k+1} (RC{k+1})")
ax[k].set_xticks(
[dh for dh in dh_range if dh % 10 == 0],
labels=[dh if dh % 50 == 0 else '' for dh in dh_range if dh % 10 == 0],
)
ax[k].set_yticks(
np.arange(0, ax[k].get_yticks().max(), 0.1),
labels=np.round(np.arange(0, ax[k].get_yticks().max(), 0.1), 2),
)
ax[k].set_ylabel("Relative Value")
if k not in r_list:
ax[k].set_xticklabels([])
ax[0].legend(loc="upper left")
ax[-1].set_xlabel("Residue Number")
ax[-2].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
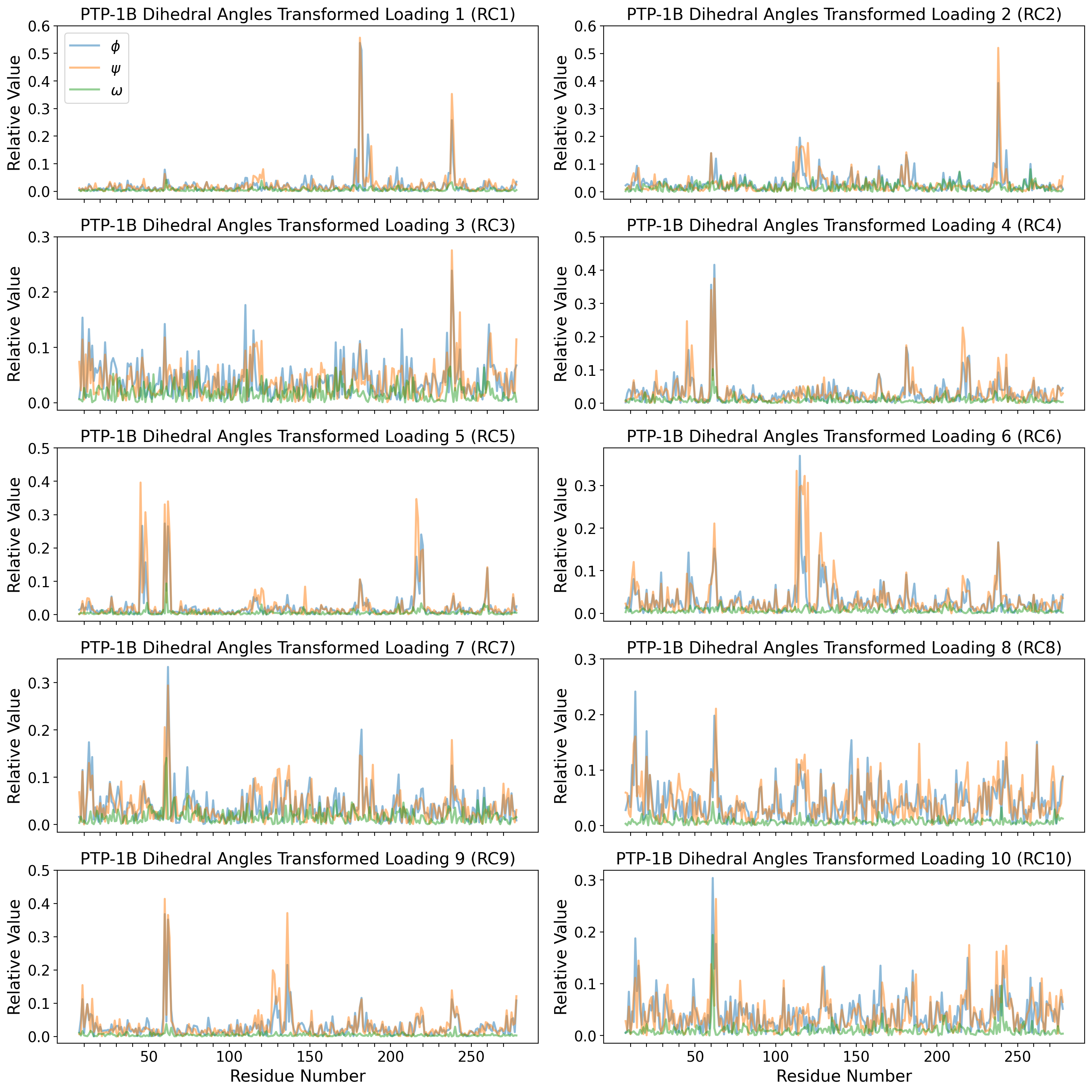
[57]:
conformations = list()
for k in pw_strucs:
if k.startswith("ptp1b_data/"):
conformations.append(conformation_dict[k.split("/")[-1][8:-4]])
else:
conformations.append(conformation_dict[k])
conformations = np.array(conformations)
print(pw_strucs[np.argmax(pw_A[:,1])])
print(pw_strucs[np.argmin(pw_A[:,1])])
pc_num = 1
plt.hist(np.array([pw_A[conformations=='open_open',pc_num],
pw_A[conformations=='open_closed',pc_num],
pw_A[conformations=='closed_open',pc_num],
pw_A[conformations=='closed_closed',pc_num]], dtype='object'),
bins=15, density=False, histtype='bar', stacked=True, label=['open/open', 'open/closed', 'closed/open', 'closed/closed'])
plt.title(f'PTP-1B Pairwise Distances PC{pc_num+1}')
plt.xlabel(f'PC{pc_num+1}')
plt.ylabel('Counts')
ptp1b_data/ptp1b_theseus_data/theseus_6bai_chainA.pdb
ptp1b_data/ptp1b_theseus_data/theseus_1g1h_chainA.pdb
[57]:
Text(0, 0.5, 'Counts')
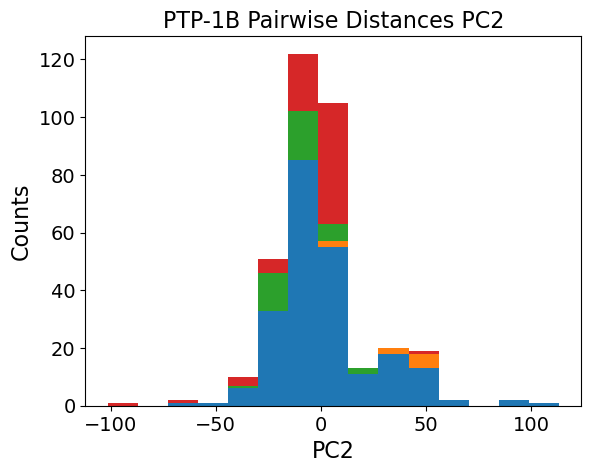
[58]:
# pairwise distances tranformed loadings
r = 10
r_list = [r - 1, r - 2]
fig, ax = plt.subplots(r // 2, 2, figsize=(15, 15), dpi=200)
ax = ax.flatten()
for k in np.arange(r):
ax[k].plot(
np.arange(resnum_bounds[0], resnum_bounds[1] + 1),
calculate_pw_rc(pw_pca.components_[k], resnum_bounds),
"-",
linewidth=2,
alpha=0.5,
)
ax[k].set_title(f"PTP-1B C\u03b1 Pairwise Distances RC{k+1}")
ax[k].set_xticks(
[x for x in np.arange(resnum_bounds[0], resnum_bounds[1] + 1) if x % 10 == 0],
labels=[
x if x % 50 == 0 else '' for x in np.arange(resnum_bounds[0], resnum_bounds[1]) if x % 10 == 0
],
)
ax[k].set_ylabel("Relative Value")
if k not in r_list:
ax[k].set_xticklabels([])
ax[-1].set_xlabel("Residue Number")
ax[-2].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
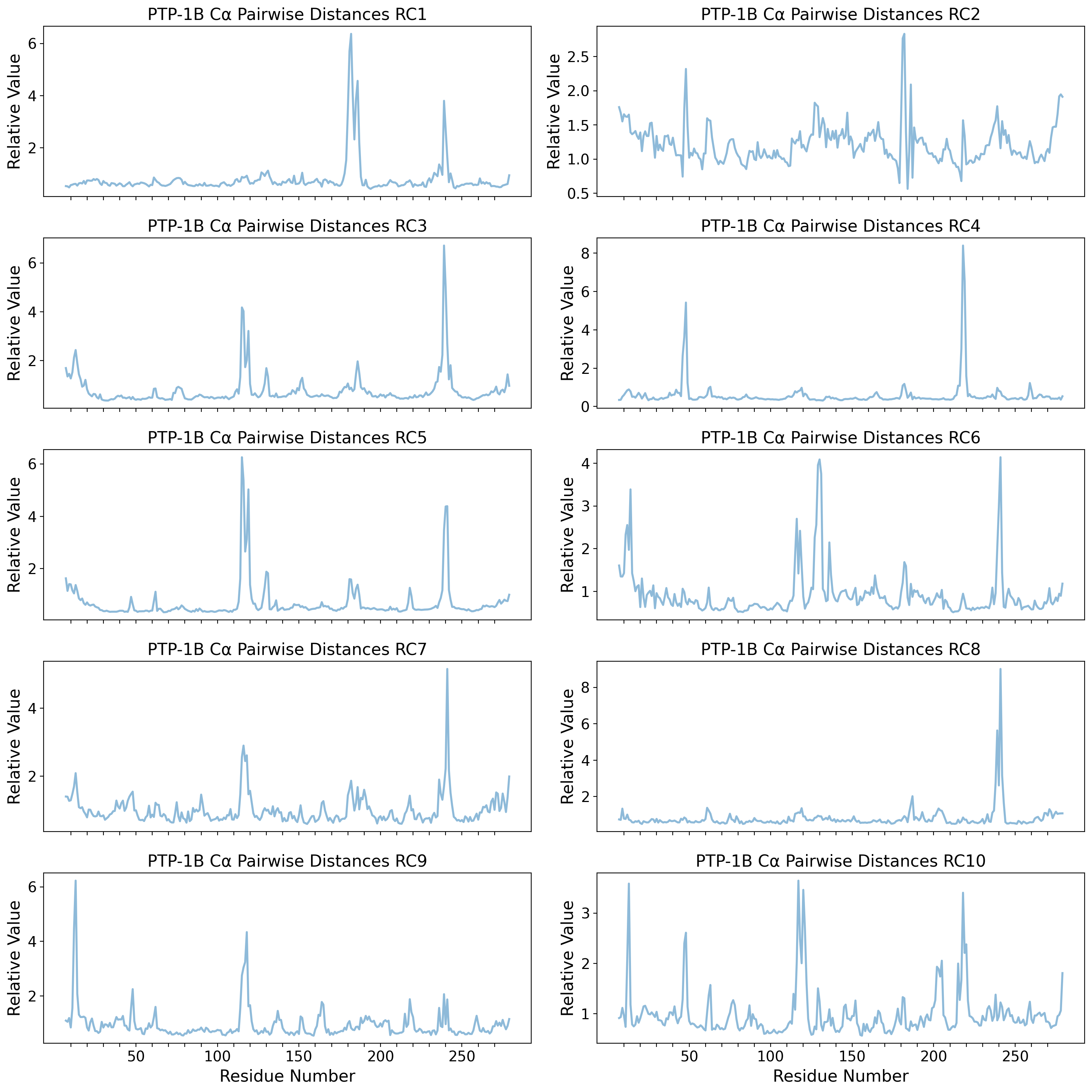
[59]:
# strain analysis tranformed loadings
r = 10
r_list = [r - 1, r - 2]
fig, ax = plt.subplots(r // 2, 2, figsize=(15, 15), dpi=200)
ax = ax.flatten()
shared_atom_set = pickle.load(open("ptp1b_data/ptp1b_atom_set.pkl", "rb"))
for k in np.arange(r):
ax[k].plot(
np.arange(resnum_bounds[0], resnum_bounds[1] + 1),
calculate_sa_rc(sa_pca.components_[k], sorted(shared_atom_set)),
"-",
linewidth=2,
alpha=0.5,
)
ax[k].set_title(f"PTP-1B Strain Analysis RC{k+1}")
ax[k].set_xticks(
[x for x in np.arange(resnum_bounds[0], resnum_bounds[1] + 1) if x % 10 == 0],
labels=[
x if x % 50 == 0 else '' for x in np.arange(resnum_bounds[0], resnum_bounds[1]) if x % 10 == 0
],
)
ax[k].set_ylabel("Relative Value")
if k not in r_list:
ax[k].set_xticklabels([])
ax[-1].set_xlabel("Residue Number")
ax[-2].set_xlabel("Residue Number")
plt.tight_layout()
plt.show()
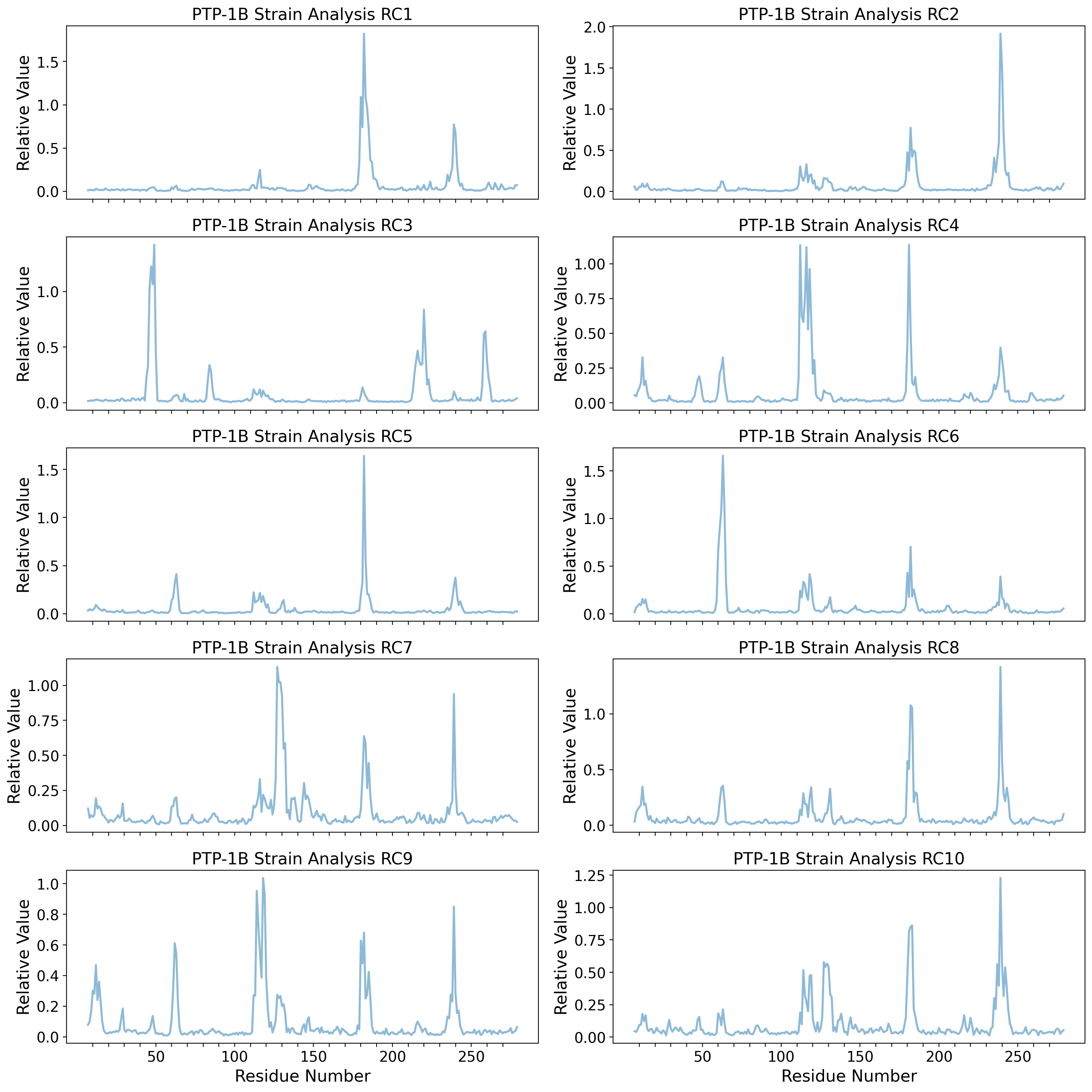
[ ]: